 © ISTOCK.COM/PEOPLEIMAGES/DEBIBISHOP
© ISTOCK.COM/PEOPLEIMAGES/DEBIBISHOP
As a postdoc at the University of California, Berkeley, in the mid-1980s, Rob Martienssen discovered a mutation in maize plants that caused their leaves to have a very pale green color. Cells carrying this mutation don’t develop chloroplasts, and as a result, the plants couldn’t perform photosynthesis. Seedlings would survive for only a week or two on stored nutrients before perishing. “This mutant had a very strong lethal phenotype,” Martienssen says. “But from time to time it reverted,” he adds. “And it reverted in a very striking way.”
Some of his mutant corn plants began to grow striped leaves, with bands of healthy dark-green tissue interspersed with the pale-green cells, and those plants survived a little longer. “From one leaf to the next, you would see more dark-green stripes,” Martienssen describes. “The plant eventually grew out of the defect altogether and flowered.”
Martienssen and his colleagues knew...
The striped-leaf pattern reminded Martienssen of the multicolored corn kernels in which famed geneticist Barbara McClintock had first identified transposons. “She had noted in the 1950s that transposable elements that landed near genes could change the expression of those genes from one generation to the next in a reversible way,” Martienssen says. “She called the phenomenon cycling, which really sums it up.” DNA methylation was a popular candidate for the control of transposon activity, and Martienssen’s work clearly supported the idea.
In 1989, when Martienssen arrived at Cold Spring Harbor Laboratory to start his own lab, McClintock was still hard at work there. She would take Martienssen out into the field and talk about the control of transposable elements in plant genomes. Both researchers were very interested in how transposons cycled on and off over subsequent generations. “She really liked the DNA methylation explanation,” Martienssen recalls.
 GENERATIONS OF EPIGENETICS RESEARCH: Famed transposon discoverer Barbara McClintock and a young Rob Martienssen inspect a maize plant at the Cold Spring Harbor Laboratory greenhouse in 1989.CSHL ARCHIVES
GENERATIONS OF EPIGENETICS RESEARCH: Famed transposon discoverer Barbara McClintock and a young Rob Martienssen inspect a maize plant at the Cold Spring Harbor Laboratory greenhouse in 1989.CSHL ARCHIVES
As the evidence mounted that methylation controlled transposon activity, plant biologists were quickly becoming convinced. But the findings were still correlational. At that time, researchers didn’t yet have the means to manipulate methylation in a controlled way to experimentally demonstrate that the epigenetic phenomenon was suppressing transposon activity. So Martienssen began mutating Arabidopsis thaliana plants, searching for one with a defect in its epigenetic machinery.
In 1993, one year after McClintock’s death, Martienssen and his colleagues found what they were looking for: the first mutant eukaryote that was defective in DNA methylation. The plants, which carried a mutation in what the researchers named the decrease in DNA methylation 1 (DDM1) gene, had significantly reduced methylation at repeated sequences in the genome, sites associated with transposable elements.2 A few years later, he and others demonstrated that transposon activation was increased in DDM1 mutant plants. “Transposons go completely crazy in terms of expression and transposon position,” explains Martienssen, who says that it was gratifying to finally have experimental evidence to support McClintock’s ideas.
In the two and a half decades since Martienssen’s brief overlap with the Nobel-winning botanist at Cold Spring Harbor, the study of plant epigenetics has exploded. In plants, DNA methylation includes not just the CG methylation that’s common in mammals and some other animals, but also CHG (where H is any base except G) and CHH methylation. And the mechanisms for maintaining these different methyl marks are diverse and intermingling. It seems that plants—whose genomes are typically large, often containing more than two sets of chromosomes, and riddled with transposons (see “Genomes Gone Wild,” The Scientist, January 2014)—are extra careful to keep much of their DNA quiet.
“It’s very important to keep these transposable elements as silenced and methylated as possible,” Martienssen says. “Because of their crazy genomes, [plants] have every possible mechanism to silence and keep things constant.”
An emerging understanding of how plants methylate their genomes could also inform our concept of animal epigenetics. For example, it was in Arabidopsis that Vincent Colot of the Institut de Biologie de l’École Normale Supérieure (IBENS) of France’s National Center for Scientific Research and colleagues identified a methylation mechanism that involves RNA interference machinery better known for its ability to suppress gene expression by degrading transcripts.3 RNA-directed DNA methylation has since been found to regulate DNA methylation at some specific loci in the mouse genome, as well as to play a role in histone methylation in Drosophila and C. elegans (and likely in mice as well, Martienssen says).
But there is a key difference in how methylation is inherited between the two kingdoms. While animal cells undergo two rounds of reprogramming during reproduction to wipe clear most of the methyl marks that decorate their DNA and histones, plants leave their epigenomes largely intact from one generation to the next. In plants, this results in epialleles—stably inherited alleles encoded by methylation, rather than by gene sequence—that control subtle phenotypes, such as timing of flowering or fruit ripening.
“Most of the differences [between individuals] that we see are caused by genetic variation,” says Colot. “But it’s not all caused by genetic variation. What would be caused by this epigenetic variation could be as important.”
Whether these epialleles can be adaptively altered by the environment remains a matter of debate, and most researchers say there is no convincing evidence for any form of such “Lamarckian” evolution. But there are hints that acquired changes in methylation patterns can impact future generations, and at the very least, errors in transcribing the methylome provide an additional source of new variation, akin to genetic mutation, as mistakes are stably inherited.
“There’s a lot more change possible epigenetically, and in some cases only possible epigenetically,” says Martienssen. “And selection just acts on that in the same way that selection would act on a genetic change.”
Deciphering the plant epigenetic code
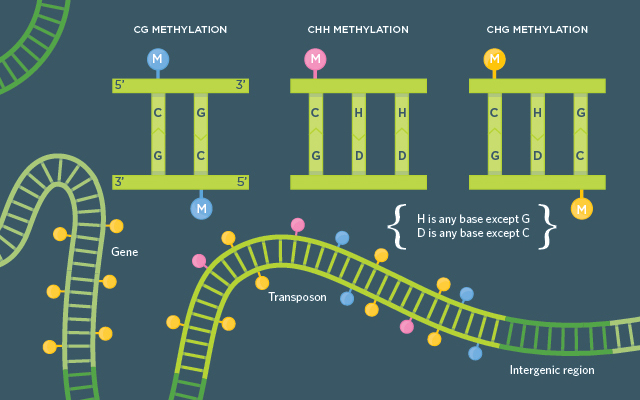 PLANT METHYLATION BASICS: In Arabidopsis, CG methylation is found on some genes, but primarily on repeat sequences that make up transposons, as well as other repeat sequences in the genome. CHH methylation is found only where there is CG methylation and often near transposable elements, though some evidence points to CHH methylation on some silenced genes as well. CHG methylation is typically found with the CHH variety.
PLANT METHYLATION BASICS: In Arabidopsis, CG methylation is found on some genes, but primarily on repeat sequences that make up transposons, as well as other repeat sequences in the genome. CHH methylation is found only where there is CG methylation and often near transposable elements, though some evidence points to CHH methylation on some silenced genes as well. CHG methylation is typically found with the CHH variety.
See full infographic: WEB | PDF© LUCY READING-IKKANDAIf there’s one technology that has revolutionized the study of epigenetics, it’s bisulfite sequencing, which converts unmethylated cytosines to uracils before DNA sequencing. In plants, it allows researchers to interrogate not just general methylation patterns—which could be deciphered by Southern blotting and microarrays— but the three specific types of methylation: CG, CHG, and CHH.
In the mid-2000s, several groups published complete methylomes of Arabidopsis. Martienssen and colleagues reported maize methylation patterns in 2013,4 and a handful of other groups followed with more maize methylomes. These surveys have revealed notable variation in methylation patterns among individuals, but unlike animals, which use epigenetics to regulate the patterns of gene expression in different cell types during development and into adulthood, plants have nearly identical patterns of methylation throughout their tissues. “For the most part, if you look at different cell types in plants, the methylation profiles are remarkably similar,” says Nathan Springer, a plant geneticist at the University of Minnesota.
This conservation of DNA methylation patterns across the plant’s tissues supports the idea that methylation is “part of a strategy to control transposable elements,” which are rampant in plants’ massive genomes, says Mary Gehring, a plant epigeneticist at the Whitehead Institute. As much as 90 percent of the maize genome, for example, is made up of transposable elements. And indeed, all three types of methylation are found on and around transposon repeat regions. “Transposons very clearly are regulated by methylation, there is no doubt,” Martienssen says.
Because of their crazy genomes, plants have every possible mechanism to silence and keep things constant.—Rob Martienssen
Cold Spring Harbor Laboratory
Methylation on genes is much less common, and when it’s there, it’s most commonly of the CG variety. But epigenetic regulation of genes may begin to crop up as researchers begin to look at other plant species, Martienssen says—in particular, those that, unlike Arabidopsis and maize, suffer lethal phenotypes when DDM1 is mutated. Last year, Bob Schmitz of the University of Georgia and colleagues greatly expanded the list of plant species with their complete methylomes mapped, surveying 34 species of flowering plants, and documented extensive epigenetic variation. Plants of the brassica family, including Arabidopsis, tend to have reduced CG methylation on genes, or none at all; grasses have more gene methylation, but of the CHH variety.5 “It became apparent, with just a few other plant species, that these rules that we’re learning from model systems don’t always apply to other species,” Schmitz says.
Whole-methylome analysis is a young technique, but more data—and hopefully more answers—are on the horizon, says Martienssen. Both Pacific Biosciences and Oxford Nanopore are “feverishly optimizing” new DNA sequencing technologies that can detect methylation without having to first swap out the methylated cytosines for uracils, he explains. “I would predict that within a year we’ll be able to sequence methylated bases just as easily as nonmethylated bases.”
Methylation inheritance
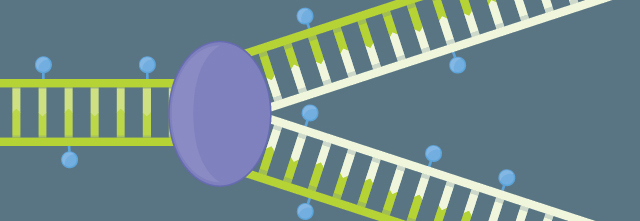 METHYLATION MAINTENANCE: Every time a cell divides, it must replicate its genome and its epigenome. Plants have diverse pathways overseeing the faithful passage of the methylome to daughter cells.
METHYLATION MAINTENANCE: Every time a cell divides, it must replicate its genome and its epigenome. Plants have diverse pathways overseeing the faithful passage of the methylome to daughter cells.
See full infographic: WEB | PDF© LUCY READING-IKKANDAAfter doing a sabbatical in Martienssen’s lab at Cold Spring Harbor, Colot returned to France in 2001 to start his own lab at the Plant Genomics Research Unit of the National Institute for Agricultural Research (INRA) with a very specific goal in mind: create epigenetic recombinant inbred lines (epiRILs) of Arabidopsis—plants with identical genomes, but different epigenomes. They could be a powerful tool for understanding the function of methylation in different parts of the genome, he reasoned, and allow researchers to track how plant methylomes are passed from one generation to the next.
Colot started with two parent plants with the same genetic background, but one parent was homozygous for a wild-type DDM1 gene, while the other was homozygous for a mutant version of the gene and thus had dramatically reduced DNA methylation. As had been suggested by previous work, the methylomes of the parents were inherited by the progeny, who each had one set of chromosomes from their wild-type parent with normal methylation patterns and a hypomethylated set from the mutant parent. Colot took one such epigenetically heterozygous female plant, bred it back to a wild-type male, and selected progeny that carried two copies of the wild-type DDM1 gene. He then selfed those plants, generating 500 separate lines with intact methylation machinery but varied methylation patterns. Even after eight or nine generations, he found that on one-third of the differentially methylated sequences, “the methylation states present in the two parents were segregated according to Mendel’s laws,” Colot said. “They are truly epigenetic”—extra-genomic variation that is faithfully passed on. The other two-thirds of the sequences he studied had regained wild-type methylation over several generations.6
This persistence of methylation patterns across the generations stands in stark contrast to what researchers knew about mammalian epigenetics, in which the CG marks are wiped out in two rounds of reprogramming in the gamete and the early embryo. For plants, this means that once methylation changes arise, they are likely to stick around. These epialleles are now known to control a variety of phenotypes, such as disease resistance, biomass, and flowering time. “There are more heritable epialleles that have been identified in plants than in mammals; presumably that’s because of this lack of reprogramming,” says Gehring. “That continues to be really exciting.”
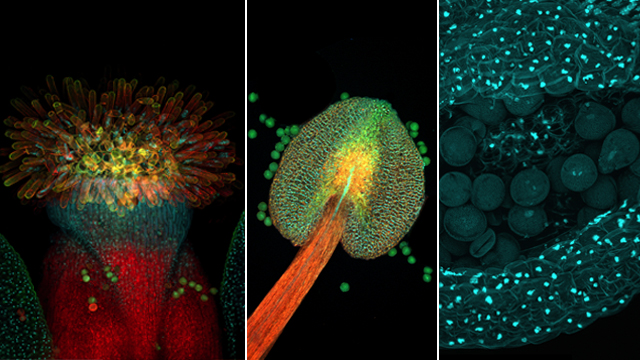 DOUBLE FERTILIZATION: In the center of an Arabidopsis flower (left), anthers (zoom shown in center image) harbor the sperm (right) that will swim through a pollen tube to fertilize the egg and the female central cell. (See illustration.)© ISTOCK.COM/HEITIPAVESBut can the environment influence the epigenome in an adaptive way? For example, if plants are exposed to a particular pathogen, could a resistance gene shed its methyl mark, improve survival, and spread as a result of Lamarckian-style evolution? Most say the evidence for it is weak. “There’s this idea that plants may be able to respond to their environment; an environmental change will affect the methylome, and then that could be inherited,” says Gehring. “There’s really not much good data that that’s true.”
DOUBLE FERTILIZATION: In the center of an Arabidopsis flower (left), anthers (zoom shown in center image) harbor the sperm (right) that will swim through a pollen tube to fertilize the egg and the female central cell. (See illustration.)© ISTOCK.COM/HEITIPAVESBut can the environment influence the epigenome in an adaptive way? For example, if plants are exposed to a particular pathogen, could a resistance gene shed its methyl mark, improve survival, and spread as a result of Lamarckian-style evolution? Most say the evidence for it is weak. “There’s this idea that plants may be able to respond to their environment; an environmental change will affect the methylome, and then that could be inherited,” says Gehring. “There’s really not much good data that that’s true.”
Others agree such evolutionary dynamics are unlikely. “There’s very little evidence that the environment perturbs DNA methylation in plant genomes,” Schmitz says. “A lot of those studies are plagued by not going out far enough. You often see [epigenetic changes in] one generation, but the second or third generation snaps back to the original generation.”
One 2016 paper aimed to address that experimental shortcoming. University of Warwick plant biologist Jose Gutierrez-Marcos and his colleagues propagated three lines of Arabidopsis, grown in normal or two grades of salty soils for five generations. From each generation, they took seeds and raised offspring and grand-offspring under normal conditions, then compared their methylomes to see whether any epigenetic changes that arose in response to salt stress persisted.7 “We used a high level of replication in our analysis. We looked at populations of individuals rather than looking at individual plants,” Gutierrez-Marcos says. “We were looking for a strong effect, not just effects that could just be spontaneously arising in the experiment.”
The researchers did see some changes in DNA methylation that were stably inherited for at least two generations. Although methylation changes began after just one generation in the salty soil, the researchers didn’t notice any phenotypic changes until the second generation, when the salt-stressed lines began to show higher germination and survival rates and more-robust foliage.
“Basically they found that not much happened,” says Gehring, still a self-proclaimed skeptic of transgenerational effects of this nature. “But they did have a couple of examples—one or two things that seemed like they might be interesting, in terms of [being] induced by stress and inherited.”
Over the five generations of the experiment, the subtle adaptive responses did not magnify, suggesting there’s a limit to epigenetic adaptation. And in the two generations spun off from each experimental cohort but raised in normal soils, some of the loci that had lost or gained methylation began to revert to the founding plant’s methylome. “But there are some particular loci that are very stable, even for four generations,” Gutierrez-Marcos noted.
If environment-induced methylome adaptation is happening in nature, it’s certainly not common, says Martienssen. “Personally I think it will be found, but obviously it will be rare. It’s not going to be a general principle.” That said, the environment doesn’t have to cause the changes for epigenetic mechanisms to be a driver of evolutionary change, he adds. “We know that epigenetic variation exists. You don’t need to have the environment directing the change so long as the changes are there.”
Schmitz says that some of this variation can be chalked up to user error. “Plants work really hard to maintain methylation, and they’re pretty good at it, but there are mistakes that occur,” he says. And because plants pass down their methylomes in a relatively stable way, “these mistakes can lead to new heritable traits.”
Reprogramming exceptions
It’s not entirely true, however, that plants do not undergo epigenetic reprogramming. During reproduction, Arabidopsis sperm don’t erase CG methylation as animals do, but they do wipe out about 90 percent of CHH methylation, which is found along with CG methylation at transposons and sometimes genes as well. In 2009, Martienssen and his colleagues published their analysis of Arabidopsis sperm methylomes, comparing the three haploid cells in pollen: two sperm cells and a vegetative cell that helps support the growth of the pollen tube that delivers the sperm to the female gametophyte housing the egg.8 Although the vegetative cell nucleus loses some CG methylation, that nucleus does not contribute genetic material to the embryo. “By way of contrast—and amazingly—the sperm cells do the opposite: they lose CHH methylation,” Martienssen says.
 PLANT SPERM SHAKE-UP: Arabidopsis sperm nuclei, which combine with the egg to form the embryo, lose CHH methylation, while the nonreproductive nuclei that contribute to endosperm formation lose some CG methylation. The reprogramming of the nonreproductive nucleus allows the exression of small RNAs that can travel to the sperm nucleus, possibly serving as guides to reinforce methylation of those transposon sequences.See full infographic: WEB | PDF
PLANT SPERM SHAKE-UP: Arabidopsis sperm nuclei, which combine with the egg to form the embryo, lose CHH methylation, while the nonreproductive nuclei that contribute to endosperm formation lose some CG methylation. The reprogramming of the nonreproductive nucleus allows the exression of small RNAs that can travel to the sperm nucleus, possibly serving as guides to reinforce methylation of those transposon sequences.See full infographic: WEB | PDF
© LUCY READING-IKKANDA
After fertilization, RNA-guided methylation pathways restore the embryo’s levels of CHH methylation. “It’s very interesting that it’s so dramatic and that it has to come back again guided by small RNAs,” Martienssen says. “It means that the small RNAs found from both parents contribute to the patterns of DNA methylation in the embryo.” Sure enough, looking at maize over subsequent generations, he and his colleagues found that the offspring of parents with differing methylomes would acquire methylation at a locus if either parent had it.4 “And we found literally thousands of places in the genome that underwent a switch that was determined by the small RNAs from either mom or dad,” Martienssen says.
The reprogramming of CG methylation in vegetative pollen nuclei is also interesting, as it results in an uptick in transposon activity that might seem detrimental to the plant’s health. But Martienssen and his colleagues speculate that there may be benefits to this strategy. For example, release of transposon silencing in the vegetative nucleus of the sperm could prompt the transcription of small RNAs, which researchers know can travel to the reproductive nuclei of the sperm and serve as guides to reinforce methylation of those transposon sequences. “If you take dead-end cell types that can never go on, why not let them express transposons, [which] get processed into small RNAs and reinforce the silencing of the transposons in the other cells around them?” Springer says of the hypothesis. “You can think of this as almost an immunization.”
Although no one has investigated epigenetic changes in plant eggs yet, there is evidence of CG reprogramming in the central cell.9 Certain nonreproductive plant tissues also undergo a form of epigenetic reprogramming. In 2015, for example, an international team of researchers found that DNA demethylation governs tomato ripening.10 And last year, plant biologist Pascal Gamas of France’s INRA and colleagues found that developing root nodules, which house symbiotic bacteria in legumes, do something similar.11 In both cases, a specific demethylase that’s only expressed in these structures removed the DNA methylation.
“Plants are really good at maintaining DNA methylation, so when you find something like this, it’s really neat,” says Schmitz. “It’s exciting because it’s so rare.”
Improving agriculture
 FLICKR, MICOLO JMany crop species are clonally propagated. In theory, this allows agriculturalists to select those individuals with the most desirable qualities—large, sweet fruits or high oil production—and grow a whole field of them. But while clonal propagation ensures the preservation of desirable genetic sequences, it doesn’t take into account the epigenome. The replacement of CHH methyl marks on the DNA in sperm, which is triggered by fertilization, is not able to take place.
FLICKR, MICOLO JMany crop species are clonally propagated. In theory, this allows agriculturalists to select those individuals with the most desirable qualities—large, sweet fruits or high oil production—and grow a whole field of them. But while clonal propagation ensures the preservation of desirable genetic sequences, it doesn’t take into account the epigenome. The replacement of CHH methyl marks on the DNA in sperm, which is triggered by fertilization, is not able to take place.
In the African oil palm, farmers noticed that between 10 percent and 20 percent of plants weren’t producing oil. In 2015, researchers learned that the poorly producing palms suffered the activation of a transposon that had lost its methylation and disrupted a gene critical for oil production.12 “[CHH methylation] is replaced inefficiently and sometimes in the wrong place,” says Martienssen, who was one of many contributors to the study. “So you get a mess.”
The loss of methylation may also unveil new phenotypes, including traits that could be advantageous to crop species. And agricultural researchers—who are often forced to cross crop plants with wild cultivars to introduce pathogen-resistance genes, for example, and then back-cross the plants to regain the desired traits (see “Putting Up Resistance,” The Scientist, June 2014)—are anxious to see what’s been hiding under all that epigenetic silencing. “If you can change the methylation, you can create heritable phenotypic variation,” Schmitz says. Any genes that are typically silenced in plant genomes are “an untapped source of diversity.”
Schmidt’s goal is to find ways to release that silencing to see if any newly expressed genes are associated with novel, potentially beneficial phenotypes. “There will be a lot of ugly-looking plants that come out of this approach,” he says. “But you just need that one that will lead to that trait of interest.”
McClintock would no doubt be impressed by the progress made in understanding DNA methylation in plants, to the point that researchers are starting to tinker with crops’ epigenomes in the hopes of making hardier or more nutritious cultivars. And if she were alive today, McClintock might well be getting her hands dirty in a similar pursuit. Even at age 90, Martienssen recalls, “she was surprisingly comfortable with modern molecular biology.” And, as it turns out, she was working on the first inquiries in the field. “Obviously they didn’t call it epigenetics in those days,” Martienssen says, “but we now know that’s what it was.”
References
- R. Martienssen et al., “Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize,” Genes Dev, 4:331-43, 1990.
- A. Vongs et al., “Arabidopsis thaliana DNA methylation mutants,” Science, 260:1926-28, 1993.
- F.K. Teixeira et al., “A role for RNAi in the selective correction of DNA methylation defects,” Science, 323:1600-04, 2009.
- M. Regulski et al., “The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA,” Genome Res, 23:1651-62, 2013.
- C.E. Niederhuth et al., “Widespread natural variation of DNA methylation within angiosperms,” Genome Biol, 17:194, 2016.
- F. Johannes et al. “Assessing the impact of transgenerational epigenetic variation on complex traits,” PLOS Genet, 5:e1000530, 2009.
- A. Wibowo et al., “Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity,” eLife, 5:e13546, 2016.
- R.K. Slotkin et al., “Epigenetic reprogramming and small RNA silencing of transposable elements in pollen,” Cell, 136:461-72, 2009.
- K. Park et al., “DNA demethylation is initiated in the central cells of Arabidopsis and rice,” PNAS, doi:10.1073/pnas.1619047114, 2016.
- R. Liu et al., “A DEMETER-like DNA demethylase governs tomato fruit ripening,” PNAS, 112:10804-09, 2015.
- C. Satgé et al., “Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula,” Nat Plants, 2:16166, 2016.
- M. Ong-Abdullah et al., “Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm,” Nature, 525:533-37, 2015.
Interested in reading more?





