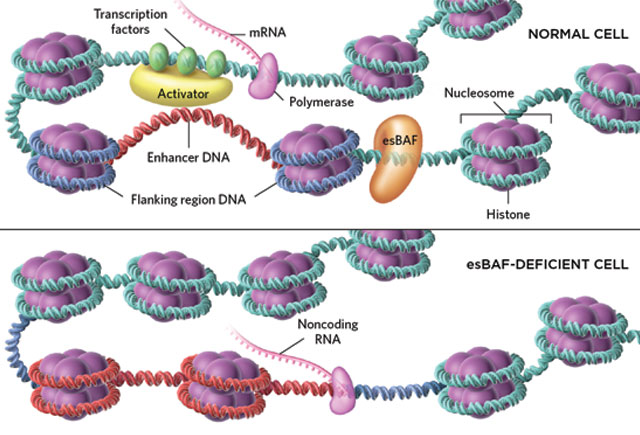
 BAF BLOCKER: In normal embryonic stem cells (top panel), the chromatin remodeling complex esBAF moves nucleosomes into enhancer-flanking DNA regions (blue) to keep enhancers (red) free of nucleosomes so they can promote transcription of protein-coding genes. When esBAF is absent (lower panel), nucleosomes move out of the flanking regions and into the enhancer, freeing up noncoding RNA transcription that begins in the enhancer and continues into the flanking DNA.© SCOTT LEIGHTON
BAF BLOCKER: In normal embryonic stem cells (top panel), the chromatin remodeling complex esBAF moves nucleosomes into enhancer-flanking DNA regions (blue) to keep enhancers (red) free of nucleosomes so they can promote transcription of protein-coding genes. When esBAF is absent (lower panel), nucleosomes move out of the flanking regions and into the enhancer, freeing up noncoding RNA transcription that begins in the enhancer and continues into the flanking DNA.© SCOTT LEIGHTON
EDITOR'S CHOICE IN GENETICS & GENOMICS
The paper
S.J. Hainer et al., “Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF,” Genes & Development, 29:362-78, 2015.
To stuff more than six feet of DNA into the nucleus of each cell in our body, the genome winds tightly around a core of eight histone proteins, creating structures called nucleosomes that, in turn, are densely packed into chromatin. To transcribe genetic information from DNA to RNA, nucleosomes...
Recent genome-sequencing work has revealed that enhancer regions not only promote the transcription of protein-coding genes, but can also contain the script for noncoding RNAs. Although noncoding RNAs are a hot topic, how they are regulated isn’t understood, says Toshio Tsukiyama of the University of Washington.
Thomas Fazzio, a former graduate student of Tsukiyama who is now an assistant professor at the University of Massachusetts Medical School, wanted to understand how the positioning of nucleosomes near enhancers might regulate noncoding RNA transcribed from the enhancer region. “We thought it was almost obvious that if BAF is important for keeping the chromatin structure of lots of enhancers open,” it would support the transcription of noncoding RNAs from those genomic stretches as well, he says. Yet when Fazzio and his colleagues sequenced the RNA from esBAF knockdown mouse embryonic stem cells, they found that noncoding RNAs from approximately 57,000 genomic regions were expressed at higher levels than in the control. The effect was not seen in more-differentiated mouse embryonic fibroblasts.
To further explore the unexpected finding, the researchers treated the DNA with an enzyme that only cuts in nucleosome-free regions and sequenced the fragments to determine the nucleosomes’ locations. Enhancer regions of esBAF knockout cells were covered by nucleosomes, but there were far fewer nucleosomes in flanking regions, suggesting that esBAF increases the nucleosome occupancy there. Adding DNA sequences that lock nucleosomes in place at both sides of the enhancer repressed the noncoding transcripts, even without esBAF. “I was really impressed that using mammalian cells they actually...directly show that it is really the nucleosome occupancy that matters for noncoding RNA regulation,” says Tsukiyama, who did not participate in the experiments.
The results point “to the intricacies of how [chromatin remodeling] complexes are used by the cell,” says Swaminathan Venkatesh of the Stowers Institute for Medical Research in Kansas City, Missouri, who was not involved in the study. “A complex that has traditionally been viewed to be involved in activating transcription is actually repressing noncoding transcripts.”
It’s not clear how important the repression of noncoding transcripts is for pluripotency, Fazzio says. There are many long noncoding RNAs implicated in differentiation, says Venkatesh, and “you can imagine that suppressing them in embryonic stem cells is preventing differentiation from taking place.”
Interested in reading more?




