 THE TECHNIQUE: A gene of interest is hybridized with a biotin-tagged DNA probe (red). Next, an anti-biotin antibody (pink) and an antibody (blue) recognizing an epigenetic mark—e.g., histone H3 methylation—are applied. These antibodies are then each tagged with PLA antibodies (orange and yellow). If the biotin and epigenetic mark are in close proximity, the two PLA antibodies will interact and create a signal detectable with a fluorescent DNA probe.GEORGE RETSECK
THE TECHNIQUE: A gene of interest is hybridized with a biotin-tagged DNA probe (red). Next, an anti-biotin antibody (pink) and an antibody (blue) recognizing an epigenetic mark—e.g., histone H3 methylation—are applied. These antibodies are then each tagged with PLA antibodies (orange and yellow). If the biotin and epigenetic mark are in close proximity, the two PLA antibodies will interact and create a signal detectable with a fluorescent DNA probe.GEORGE RETSECK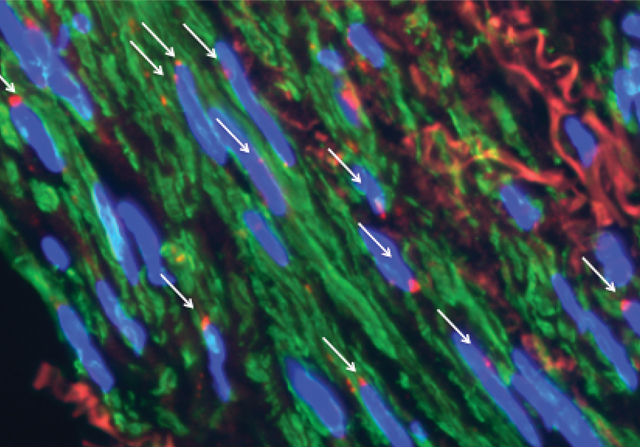 THE PICTURE: Red fluorescent signals (white arrows) appear only in nuclei (blue) where the biotinylated gene probe and epigenetic mark are in close proximity.IMAGE COURTESY OF DELPHINE GOMEZ
THE PICTURE: Red fluorescent signals (white arrows) appear only in nuclei (blue) where the biotinylated gene probe and epigenetic mark are in close proximity.IMAGE COURTESY OF DELPHINE GOMEZ
Techniques exist to visualize specific gene loci within tissue sections. And separate test-tube experiments exist to determine those genes’ epigenetic modifications. Now Gary Owens, a professor of cardiovascular research at the University of Virginia, has devised a new technique that enables gene visualization and epigenetic analysis at the same...
“The dirty little secret of epigenetics research is that we report quantitative differences from a cell population,” says Andrew Feinberg, a professor of molecular medicine at Johns Hopkins University who was not involved in the study. “If you really want to understand mechanisms, you also need to measure individual cells.”
To achieve single-cell precision, Owens modified an existing technique called a proximity ligation assay (PLA) that is used to determine if two proteins are in close proximity within a cell. Antibodies to the two proteins are tagged with overlapping complementary single strands of DNA. If the two strands are close together, they can be ligated to form an amplifiable circular DNA molecule that is detectable with a fluorescently labeled DNA probe.
Owens simply combined the method with in situ hybridization (ISH) to reveal whether a specific gene was in close proximity to a particular epigenetic mark—for example, methylation of lysine 4 in histone H3. First he hybridized his gene of interest with a DNA probe tagged with biotin. He then used an anti-biotin antibody tagged with one of the single-stranded DNAs for PLA. The other PLA tag was attached to the antibody recognizing the particular epigenetic mark.
The concept of combining ISH with PLA “was a real technical tour de force,” says Feinberg.
At present, Owens’s PLA-ISH technique allows examination of one type of epigenetic mark at one gene locus per experiment. But he says that with the development of further PLA reagents, it should be possible to look at multiple loci, epigenetic marks, and even transcription factors at the same time. (Nat Methods, 10:171-77, 2013)
![]()
| THE METHOD | HOW IT WORKS | ADVANTAGES | DISADVANTAGES |
| Chromatin immunoprecipitation (ChIP) | DNA cross-linked to chromatin is sheared into short sections. Antibodies recognizing epigenetic marks precipitate the associated DNA segments, which can then be sequenced. | Robust, well-established | Results are an average from pooled cells. |
| PLA-ISH | A gene of interest is tagged with single stranded DNA (ssDNA), and modified histones are tagged with a complementary strand of ssDNA. If the gene and modified histones are in close proximity, the ssDNAs anneal and are detected with a fluorescent probe. | Precise detection of specific proteins at specific gene loci in tissue sections | Currently limited to one color/interaction type per experiment |
Interested in reading more?





