However, most NGS library prep kits fail to adequately capture RNAs of a diverse range of sizes from limited and low-quality samples, such as formalin-fixed, paraffin-embedded (FFPE) samples. This is due to the efficiency of the enzyme that is responsible for adding adapters required for library amplification. Specifically, reactions are prone to nucleotide bias – some fragments with certain nucleotides at the 3’ end will be ligated with better efficiency than other fragments, resulting in a biased library.
Consequently, researchers who want to profile both small and large RNAs need to create two separate libraries. One library would contain only long RNAs, and the other would contain only short ones, while RNAs of intermediate size would get lost altogether. Each library would need to be sequenced separately and then researchers would need to use bioinformatics to normalize gene expression between the two datasets. This process is not only time consuming,...
The SEQuoia Complete Stranded RNA Library Prep Kit from Bio-Rad Laboratories captures a greater diversity of small RNAs compared to other commercial kits, including snoRNA, miRNA, tRNA, and snRNA (Figure 1). The efficient, simultaneous capture of diverse RNA subtypes results in a library that more accurately represents the complete transcriptome and allows researchers to discover biologically relevant biomarkers beyond mRNA.
The kit is compatible with a broad range of input RNA quantities (Figure 2) yieldling accurate sequencing reads with as little as 100 pg of RNA.
The Secret is in the Enzyme
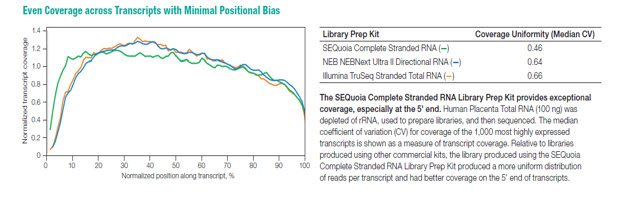
The kit features SEQzyme, a proprietary enzyme that couples cDNA synthesis and the addition of adaptors in a continuous synthesis reaction. Bio-Rad’s data reveal that the SEQuoia kit can produce greater library yields with fewer PCR cycles, even when low quantities of degraded RNA is used. (Figure 3). SEQuoia libraries generated from FFPE samples also exhibit a higher concordance rate with libraries from frozen samples when compared to other commercial kits (Figure 4). Transcript coverage was also more uniform over the entire transcript (Figure 5)
Bio-Rad’s workflow minimizes the number of pipetting steps and reduces the protocol time to under four hours, which is less than two-thirds the time of competitor protocols. Plus, this kit enables the construction of RNA libraries that are more than 99% stranded and suitable for next-generation sequencing on Illumina® platforms.
Looking Forward
After sequencing the human genome, scientists realized that analyzing gene expression by sequencing the transcriptome would bring us even closer to discovering biomarkers of disease. The challenges in capturing small RNAs for NGS library preparation have made it difficult to address the relationship between gene expression and disease and establish a foundation for drug discovery. The SEQuoia Complete Stranded RNA Library Prep Kit, with its consistency
across a range of input RNA quantities, and the ability to generate rich data sets even from challenging samples, should bring scientists closer to solving the genetic basis of disease.
Figure 1:
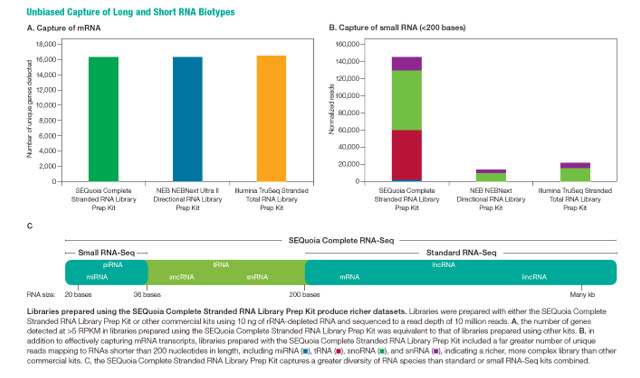
Figure 2:
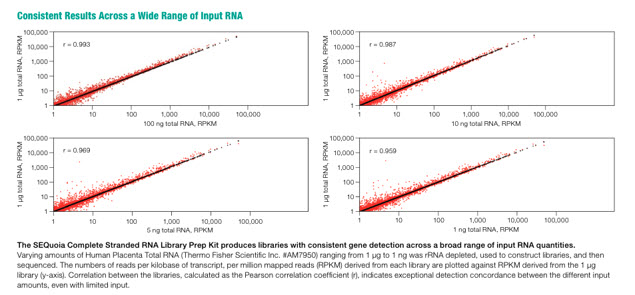
Figure 3:
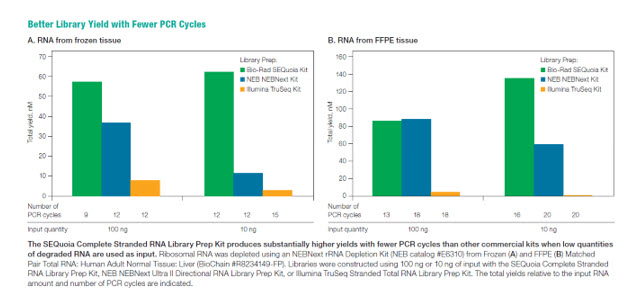
Figure 4:
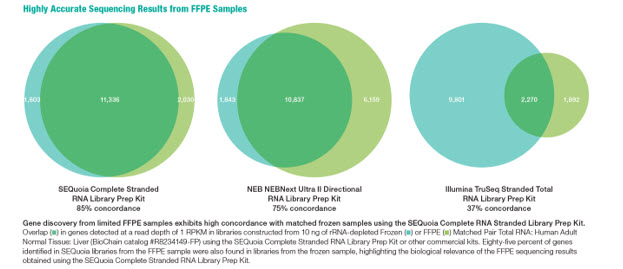
Figure 5:
Interested in reading more?




