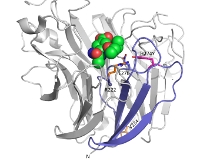
 |
with the resistance mutation (H274Y) shown in pink. The two compensatory mutations (V234M and R222Q) at the sites shown in orange may help the protein folding in a way that rescues viral fitness in the presence of the H274Y mutation. (The figure was generated using the atomic coordinates from a crystal structure (PDB ID. 3CL0) on Pymol.) Image: Kalyan Das (kalyan@cabm.rutgers.edu) CABM & Rutgers University, NJ |
**__Related stories:__***linkurl:Flu clues;http://www.the-scientist.com/article/display/57243/
[April 2010]*linkurl:Flu-drug flap;http://www.the-scientist.com/news/display/56141/
[10th November 2009]*linkurl:Single-handed flu combat?;http://www.the-scientist.com/blog/display/55443/
[23rd February 2009]














