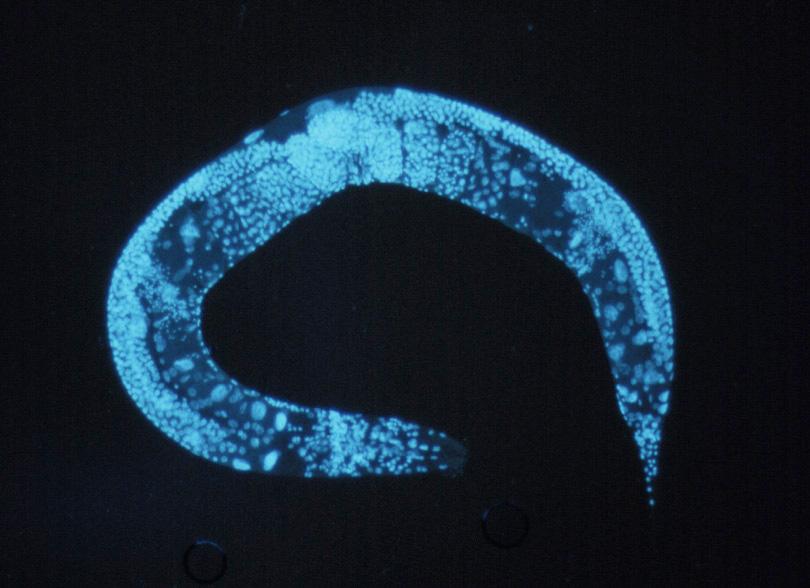
 |
Image: Wikimedia commons |
 |
Image: Wikimedia commons |
**__Related stories:__***linkurl:Supermodels?;http://www.the-scientist.com/blog/display/57225/
[19th March 2010]*linkurl:First pages of regulation;http://www.the-scientist.com/news/display/53280/
[13th June 2007]*linkurl:Post-genome project launches;http://www.the-scientist.com/news/20030305/02/
[5th March 2003]


















