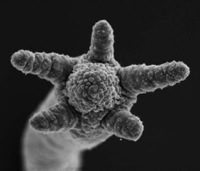
Hydra tentacle formation during budding
Bosch's team scanned all the messenger RNAs of two closely related __Hydra__ species for genes differentially expressed in the main polyp-specific structures -- tentacles, nematocysts, and the stalk. Their transcriptome-tracking turned up __Hym301__, a gene coding for a secreted protein that was expressed in the tentacles of one species and everywhere but the tentacles in the other species. By using transgenic and mutant Hydra that overexpressed __Hym301__, as well as RNA interference to silence __Hym301__, they showed that the gene affects the general timing and order in which tentacles arise in the different linkurl:cnidarians.;http://www.the-scientist.com/news/display/53364 Genes such as __Hym301__ that don't resemble known coding sequences in any other organisms -- known as "orphan" genes -- are thought to constitute around 5 to 10% of all genes across most taxonomic ranks, and, yet, geneticists have largely ignored them for two reasons. First, characterizing a gene of no known function is time-consuming and laborious. Second, many suspected that these genes had counterparts in other organisms, but limited data precluded their detection. "You can't ignore [orphan genes] anymore because databases are getting very complete," Bosch told __The Scientist__. "There are obviously evolutionary selective constraints on keeping them for millions of years... They can't just be nonsense genes that are lying around." Perhaps the best examples of orphan genes, noted Bosch, can be found in the innate immune system where organisms need highly self-specific defense mechanisms. He said he also has unpublished results of __Hydra__ anti-microbial peptide genes with no genetic equivalents outside the genus. Further, he points to human beta-defensin as another lineage-restricted antimicrobial peptide gene that is found only in mammals. "These novel genes are important for adapting an organism to its particular needs," Bosch said. "That's why these genes are not found outside [their] taxon." Caltech evo-devo biologist linkurl:Eric Davidson,;http://www.its.caltech.edu/~davidson/ who was not involved in the study, however, doesn't think that the results of the paper can be generalized to account for fundamental evolutionary processes in other organisms. "What makes [a] body plan is fundamentally and generally the deployment of regulatory genes, not the specialized downstream genes," he wrote in an E-mail. Wagner takes a more nuanced stance toward the study's implications, though. "It certainly doesn't undermine the fact that cis-regulatory changes are important in morphological evolution, but it broadens the horizon by showing that other mechanisms, including new genes, can contribute to morphological differences." Image courtesy of PLoS Biology















