 |
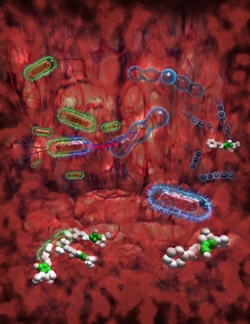
ecology in the gut. Pathogenic bacteria (green coats) receiving Penicillin resistance genes from beneficial gut bacteria (blue rounded chains) Image courtesy of A. Canossa, M. Sommer and G. Dantas |
**__Related stories:__***linkurl:The Microbial Health Factor;http://www.the-scientist.com/article/display/55864/
[August 2009]*linkurl:The number two-ome;http://www.the-scientist.com/article/display/55786/
[July 2009]*linkurl:Mysterious resistance;http://www.the-scientist.com/article/display/54963/
[September 2008]




















