Over 100 years ago, German neurologist Korbinian Brodmann stained post-mortem samples of the human cerebral cortex and observed their architecture under a light microscope.1 He drew borders based on regional variations in cell shape, density, and size, and came up with 52 distinct regions, many of which applied to the brains of other mammals as well. Since then, the rise in use of animal models, whole-brain imaging techniques, and single-cell analysis methods like spatial transcriptomics has enabled scientists to produce brain maps with much more detail. One of the latest 3D atlases of the mouse brain, the Allen Mouse Brain Common Coordinate Framework version 3 (CCFv3), consists of over 600 regions.2
While such advances bring with them a breadth of knowledge, the immense complexity of the datasets makes it challenging for scientists to analyze them using current computational methods. Now, researchers at the Allen Institute for Brain Science and the University of California, San Francisco (UCSF) have created an artificial intelligence (AI) model called CellTransformer that can automatically sort cells into subregions in the mouse brain based on spatial transcriptomics data from millions of dissociated cells.3 Using this advanced neural network, the team created one of the most detailed maps of the mouse brain to date, featuring 1,300 regions. Their findings, published in Nature Communications, could enable scientists and clinicians to study how different neurons link to behaviors and diseases at a finer scale.
“There could be several questions you could answer with this method,” said Per Uhlén, a neuroscientist at the Karolinska Institute who was not involved in the study. “If you look into different mutations that are known [to] cause diseases, maybe those are restricted to a certain region that you didn't know existed. With this method, you could localize where in the brain these mutated cells reside.”
To build CellTransformer, Bosiljka Tasic, a neurobiologist at the Allen Institute for Brain Science, Reza Abbasi-Asl, a computational neuroscientist at UCSF, and their team used a dataset comprised of 3.9 million cells from mouse brains categorized into more than 5,000 types of neurons. Each cell was analyzed for 500 different genes.
“We thought that this rich data could help us define brain regions in a data-driven way. But how do you do that?” Tasic said. “How do you do it systematically without saying, ‘Oh, I see this cell type here mostly, so I'm going to draw a boundary where that cell type is absent.’ How do you do it for all the cell types?”
Instead of relying on visually discernible boundaries, the team designed the algorithm to define the context of each cell type based on the gene expression data and compare it to the context of other cells, similar to how ChatGPT assesses words in a sentence. “You can imagine how similar are words in the language based on the context that they appear,” Tasic said. “Here we are asking how similar cells are based on the context in which they appear. And then once we define these neighborhoods for each cell, we can cluster them together.”
Once the researchers had the model ready to go, they experimented with how many regions they divide the mouse brain into. With more conservative inputs, CellTransformer generated maps that matched well with the established atlases. But Tasic and her team wanted to see how fine a division they could get with lowering the constraints in the tool. “I like to see all the differences that are there, even if they’re not at the super high statistical significance. But I want to see it. I want to describe it. And I want to put it out to the world and then ask if anybody else finds an orthogonal way of finding that same division,” Tasic said.
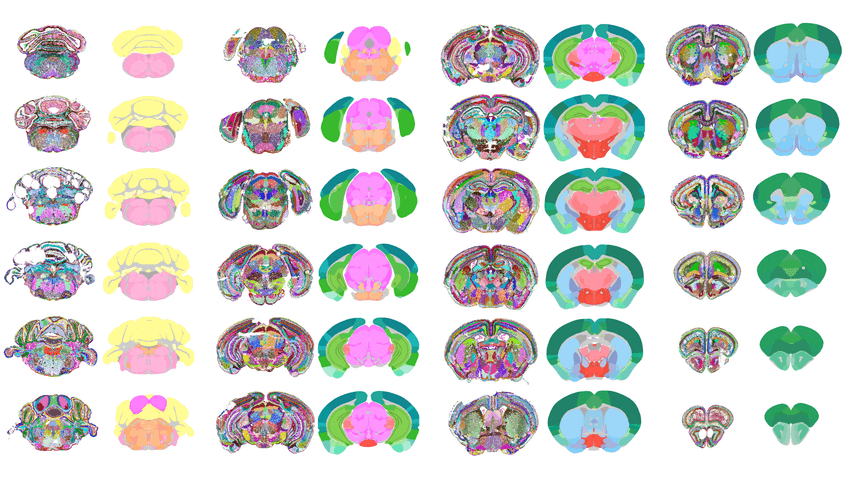
Researchers at the Allen Institute of Brain Science and the University of California, San Francisco created a new map of the mouse brain, featuring 1,300 regions/subregions. Shown here are a few examples.
University of California, San Francisco
With this approach, they discovered that the mouse brain can be divided into 1,300 distinct regions or subregions, some of which scientists have never observed before. They tested the model on different datasets, with variations in the planes of brain dissection, transcriptomics methods, and resolution of sampling. They could generate a map with the same level of detail using them all. “It surprised me that they could find so many more subregions. That is very interesting,” Uhlén said. “Maybe not all these subregions are relevant. Maybe some of them are. This remains to be tested.” He added that experiments in living tissue are essential to determine if these subregions are indeed functionally distinct.
Tasic and her colleagues are now applying the model to other kinds of data, such as neural projections and activity patterns in the brain, to test if they observe the same divisions.
“I think it could be applied to any organ, any tissue. The hope is that it can be applied to even bigger tissues like those of primates or humans. But we still need to collect a lot of data,” Tasic said.
- Zilles K. Brodmann: A pioneer of human brain mapping—his impact on concepts of cortical organization. Brain. 2018;141(11):3262-3278.
- Wang Q, et al. The Allen mouse brain common coordinate framework: A 3D reference atlas. Cell. 2020;181(4):936-953.e20.
- Lee AJ, et al. Data-driven fine-grained region discovery in the mouse brain with transformers. Nat Comm. 2024.















