 |
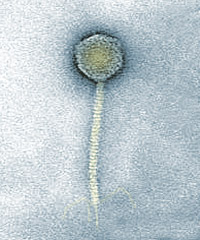
bacteria such as Lambda phage Image:Flickr/ACJ1 |
**__Related stories:__***linkurl:Package delivery;http://www.the-scientist.com/article/display/57164/
[March 2010]*linkurl:Bacterial genes jump to host;http://www.the-scientist.com/news/display/53552/
[30th August 2007]*linkurl:A new breed of viral invasion;http://www.the-scientist.com/blog/display/56239/
[6th January 2010]


















