Circular RNAs often form as the pre-mRNA molecule is processed into its final transcript via splicing, in which introns are removed and exons are linked together. Most circular RNAs are thought to be formed by a process called backsplicing, which joins one end of an exon to the other, or to an upstream exon, forming a circle. Researchers have recently published several models—not all of them necessarily mutually exclusive—to explain how different parts of the RNA molecule are brought into close proximity, encouraging backsplicing and turning a linear sequence into circular RNA.
 THE SCIENTIST STAFF
THE SCIENTIST STAFF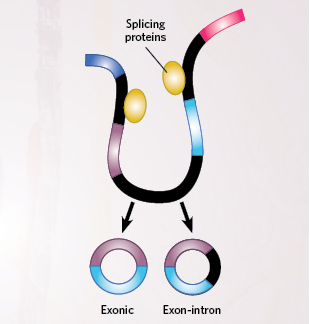 THE SCIENTIST STAFFIn a general backsplicing model, proteins assemble to form the spliceosome that processes transcribed RNA. But instead of splicing exons together in a linear sequence, they join the end of one exon to the beginning of the same exon or to an upstream exon. Below are three mechanisms that can drive this backsplicing
THE SCIENTIST STAFFIn a general backsplicing model, proteins assemble to form the spliceosome that processes transcribed RNA. But instead of splicing exons together in a linear sequence, they join the end of one exon to the beginning of the same exon or to an upstream exon. Below are three mechanisms that can drive this backsplicing
INTRON-PAIRING-DRIVEN CIRCULARIZATION
Complementary base pairs formed between long intronic sequences on different parts of the transcript bring together different splice sites on an RNA molecule, promoting backsplicing.
LARIAT-DRIVEN CIRCULARIZATION
Splicing proteins “skip” some ...













