
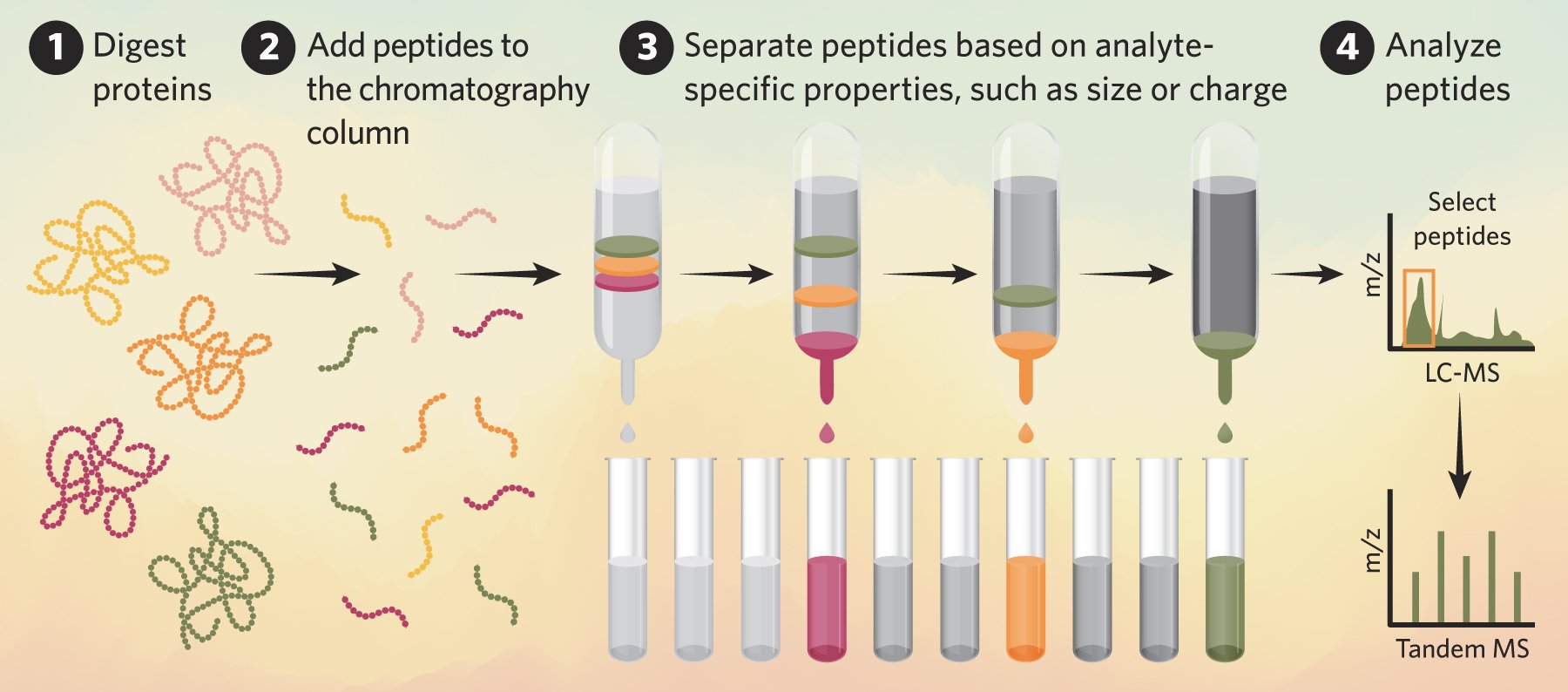
Researchers collect LC-MS data as spectrographs that show the mass-to-charge ratio (m/z) of different analytes in a sample.
Stay up to date on the latest science with Brush Up Summaries.
What Is Liquid Chromatography-Mass Spectrometry?
Liquid chromatography-mass spectrometry (LC-MS) is a laboratory technique that combines two analytical processes to separate, identify, and measure molecules in a liquid sample. Researchers use LC-MS, also called peptide mass fingerprinting, and variations of this technique to determine the amount of different analytes in a sample, including proteins, metabolites, and therapeutics.1,2 In clinical laboratories,3 scientists often perform two MS steps for tandem MS (LC-MS/MS), which increases analysis specificity compared to single stage LC-MS.2
How Does LC-MS Work?
Scientists isolate a sample’s components with liquid chromatography (LC), which separates molecules based on how they interact with the mobile and stationary phases in a column.4 These interactions depend on analyte properties, such as charge and size. Once scientists separate the individual components, the sample is further analyzed with mass spectrometry (MS), where a machine measures and reports the molecular weight and ionic charge of components in the sample. This allows researchers to ascertain which components are in a sample and quantify the amount of each analyte.1
Most researchers report LC-MS data as a mass-to-charge ratio (m/z), where m denotes the molecular weight of the ion and z represents the number of charges.4 For small molecules with a single charge, the m/z value is the same as the mass of the molecular ion, whereas larger molecules such as proteins or peptides carry multiple ionic charges and have unique m/z ratios that are fractions of their molecular weight.4 LC-MS enables molecular weight determination, which provides scientists with insight into how components in a sample are modified or whether their sample contains the expected analytes.5
LC-MS Applications
Proteomics
Proteomic analyses allow scientists to characterize the entire protein content in a sample. This may include identifying which proteins are in a sample, whether they are regulated by post-translational modifications such as phosphorylation or glycosylation, and which conformations or interactions the proteins form.6
Scientists often employ a bottom-up approach to proteomics, which is an analytical workflow that relies on peptide sequences rather than intact proteins. Researchers first digest the proteins into smaller peptides with proteolytic enzymes such as trypsin. They then use LC-MS/MS to isolate and identify these peptides and reassign the peptide sequences to the originating proteins through a process called protein inference. Scientists use this approach to uncover biological processes in cells and tissues, and to identify new drug targets and diagnostics tools through protein biomarker discovery.6
Metabolomics
Researchers comprehensively profile metabolites through metabolomics, which serves as a direct functional readout for many biological processes. Among the metabolomics technical platforms that scientists turn to, untargeted LC/MS enables researchers to measure thousands of metabolites simultaneously.7 A typical untargeted metabolomics workflow identifies features of unknown metabolites by retention time in LC and the mass-to-charge ratio in MS. However, untargeted metabolomics may also unintentionally measure molecules that are not metabolic molecules. Scientists rely on tandem MS to examine specific metabolite features in detail, or coelute isotopically labeled reference samples during LC to ensure they are studying true metabolic markers.7
Additionally, with the advent of high-resolution spectrometers, LC-HRMS (liquid chromatography-high-resolution mass spectrometry) has become the analytical tool of choice for metabolomics.8 With high sensitivity, simple sample preparation, and broad small molecule coverage,8 LC-HRMS overcomes throughput limitations of conventional LC-MS/MS for large-scale metabolomics.7
Pharmacokinetics and drug discovery
Scientists use LC-MS to analyze pharmaceuticals and differentiate the molecules that make up drugs and their byproducts.4 LC-MS enables fast and accurate quantitative measurements, which are important in drug toxicology for clinical studies, impurity profiling, doping control analysis, food sciences, and environmental research such as water analysis.9
MS-based measurements are also useful in pharmacokinetic studies, drug discovery, and biopharmaceutical research. Researchers employ MS in more than 95 percent of pharmaceutical product quantitation studies.9

- National Cancer Institute. Liquid chromatography–mass spectrometry. In: NCI Dictionaries. Accessed July 11, 2023.
- Pitt JJ. Principles and applications of liquid chromatography-mass spectrometry in clinical biochemistry. Clin Biochem Rev. 2009;30(1):19-34.
- Grebe SK, Singh RJ. LC-MS/MS in the clinical laboratory – where to from here. Clin Biochem Rev. 2011;32(1):5-31.
- Garg E, Zubair M. Mass Spectometer. In: StatPearls [Internet]. 2023. Accessed July 10, 2023.
- Lippens JL, et al. Rapid LC-MS method for accurate molecular weight determination of membrane and hydrophobic proteins. Anal Chem. 2018;90(22):13616-23.
- Gillet LC, et al. Mass spectrometry applied to bottom-up proteomics: entering the high-throughput era for hypothesis testing. Annu Rev Anal Chem (Palo Alto Calif). 2016;9(1):449-72.
- Li S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123.
- Schiffman C, et al. Filtering procedures for untargeted LC-MS metabolomics data. BMC Bioinformatics. 2019;20(1):334.
- Loos G, et al. Quantitative mass spectrometry methods for pharmaceutical analysis. Philos Trans A Math Phys Eng Sci. 2016;374(2079):20150366.












