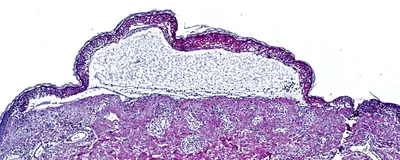
During his training in clinical dermatology, Thierry Nordmann encountered several patients with a rare, horrifying condition called toxic epidermal necrolysis, or TEN. Widely considered to be the only emergency in the field of dermatology, TEN has no effective treatments and is often fatal. “I experienced firsthand what it means not to have specific treatments for this disease, and what that means for the patient,” said Nordmann, a clinician scientist at the Max Planck Institute of Biochemistry. “For me, it was really an eye opener in the sense of this is the disease I want to study.”
He had his work cut out for him—TEN is rare, and the rapid progression makes it difficult to study. But through the use of the innovative spatial analysis technique, deep visual proteomics (DVP), a TEN treatment may finally be on the horizon.
TEN occurs when patients have severe immune reactions to certain drugs, such as antibiotics; the rapid-onset condition causes the death of keratinocytes, resulting in fluid-filled blisters and detachment of the epidermis over more than 30 percent of the body.1 “Your skin starts to peel off, and you will basically die from this,” explained Falk Schlaudraff, a molecular biologist and senior research scientist at Leica Microsystems. “We are talking about a reaction which takes place within hours, so you don't have a lot of time—you basically need to come directly to the emergency hospital to survive.”

During his clinical dermatology training, clinician scientist Thierry Nordmann was unable to treat patients with toxic epidermal necrolysis. Using DVP, he has identified both the cause of the disease as well as a potential cure.
Susanne Vondenbusch-Teetz, MPI of Biochemistry
At the time that Nordmann began studying TEN, no one knew the molecular mechanisms that caused it or why some people developed only a mild relative of the disease, known as Stevens-Johnson syndrome (SJS).
Nordmann was inspired by the work of Matthias Mann, a pioneer in DVP at the Max Planck Institute of Biochemistry. He wrote asking if there were research positions available in Mann’s lab and explained that he already had patient samples from his time in the clinic, formalin-fixed and embedded in paraffin blocks. He got the job and set out to study TEN with the goal of providing treatment options for patients.
Since so many previous studies had fallen short of the mark, Nordmann knew he would need a completely different approach if he was to identify the key drivers of the disease. “To be honest, I [thought], ‘I'm going to take a whole new look at this, do something that has not been done before, and see what comes out’,” he said. DVP provided the answer. This method capitalizes on recent scientific and technological advances to map the proteomes of individual cells or nuclei within a formalin-fixed, paraffin-embedded (FFPE) tissue section while retaining their original spatial context.
In a typical DVP workflow, researchers use high-resolution microscopy to identify cells of interest, artificial intelligence-based image analysis for cell segmentation, laser-capture microdissection to excise the cells from the tissue section, and ultra-high sensitivity mass spectrometry to analyze the proteome of those cells. Finally, they can map the data back to the original tissue section to understand their spatial context—where they sit within the layers of the skin dermis, for example.
For their samples, Nordmann and Mann needed a high-throughput approach, leading them to initiate a collaboration with Leica Microsystems, who provided laser-capture microdissection instrumentation and software. One of the key modifications involved delivering dissected cells directly into multi-well plates, allowing the team to perform more high-throughput downstream analysis.
Nordmann, Mann, and their colleagues developed a protocol for fluorescence staining to separate the keratinocytes from the immune cells in the dermis and segment them accordingly. “This way, [Nordmann] was able to figure out what is in the proteome expressed in the keratinocytes and in the immune cells and in the different forms of the disease, so he was able to compare the mild forms versus the lethal one,” said Schlaudraff. “This is basically the beauty of DVP, that you don't need to have a hypothesis of what you are looking for.”
Back in Mann’s lab, Nordmann said he did a very broad analysis, comparing the skin cells and skin-infiltrating immune cells in samples of TEN with SJS, healthy skin, and other types of drug-induced skin reactions. Nordmann said the findings, published in a study in Nature, were unusually striking: All TEN patients had drastically elevated interferon signatures—including the JAK/STAT pathway—in their cells.2
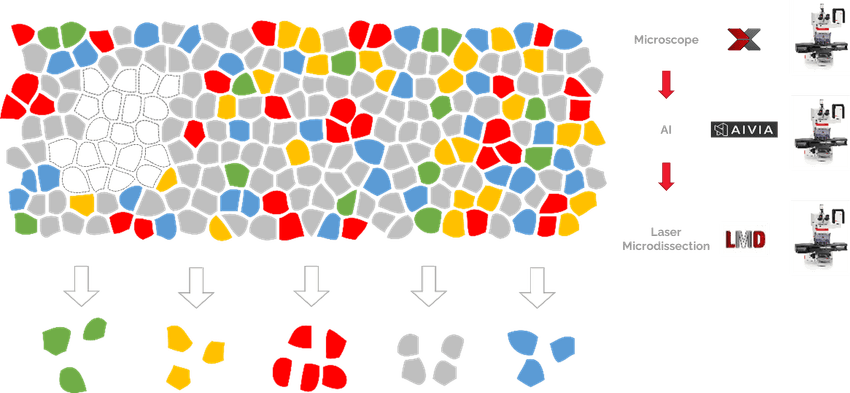
In DVP workflows, laser-capture microdissection allows scientists to map the proteome of cells of interest back to their original tissue for spatial context.
Leica Microsystems
“Usually, you get a lot of heterogeneity between patients, you know, you get mixed signals. But it was so clear that this signature was an interferon signature in all patients that we studied in comparison to both healthy and other drug reactions,” Nordmann remarked. “Once you saw this plot of these proteins that were highlighted in this disease, it was a magical moment.”
The magic didn’t stop there: Every subsequent experiment confirmed the initial results. There was no good in vitro model for TEN at the time, so the team created their own; taking keratinocytes from real TEN patients, the team grew the cells in vitro and then mixed them with immune cells from the same patients to successfully mimic the disorder. Fortunately, there are already approved drugs that can target other types of interferon-driven diseases, such as JAK inhibitors (JAKi). The team immediately demonstrated the efficacy of JAKi in their in vitro model.
The next step was to demonstrate efficacy in an in vivo model, Nordmann said. “We thought, ‘Okay, we can't stop there because that's not going to help the patient’.” With the help of more international collaborators, they tested JAKi in two distinct mouse models of the disease, including a humanized mouse model. Again, the team were encouraged by the robustness of the results. “Every single mouse model we tested, and every single experiment, was perfection. There was not one outlier,” Nordmann said.
The JAKi tested by the team have not yet been approved for the treatment of TEN, but Nordmann said he and his colleagues are pushing hard towards initiating a randomized clinical trial for the disease. “We're strategically aligning with [pharmaceutical companies] to see how do we best approach this and designing this study very carefully with the proper endpoints so that we can address this,” said Nordmann. “This will still take time. I think, optimistically, we will be there in three to five years.”
In the meantime, both Nordmann and Schlaudraff are continuing to improve DVP methods and apply the powerful technique in the study of other diseases. Nordmann has now founded his own laboratory at the Max Planck Institute of Biochemistry to research the molecular and spatial biology of the skin, and the team focuses on the use of DVP to systematically profile clinical specimens of skin diseases. “You can basically separate the good cells from the bad cells—and even from the emerging bad cells—and then hopefully find an interaction point which is a druggable target or biomarkers that you can [use for] early diagnosis of any kind of disease,” Schlaudraff added.
Correction: This article was updated on October 8, 2025 to correct that Nordmann, Mann, and their colleagues developed a fluorescence staining protocol to separate keratinocytes from immune cells in the skin. Schlaudraff did not help develop that protocol.
- Shah H, et al. Update on Stevens–Johnson syndrome and toxic epidermal necrolysis: Diagnosis and management. Am J Clin Dermatol. 2024;25(6):891-908.
- Nordmann TM, et al. Spatial proteomics identifies JAKi as treatment for a lethal skin disease. Nature. 2024;635(8040):1001-1009.