DNA methylation is a key epigenome component that helps dictate how genes are expressed, contributing to normal cell and tissue differentiation during development, as well as the process of biological aging. However, aberrant DNA methylation is involved in many different diseases, including cancer, neurological disorders, and autoimmune disease. In this article, explore what DNA methylation is and how it affects gene expression, techniques for DNA methylation sequencing, and the effect of DNA methylation on human health.

DNA methylation controls gene expression without altering the underlying DNA sequence.
iStock, agsandrew
What Is DNA Methylation?
DNA methylation is a type of epigenetic DNA modification that affects gene expression without changing the underlying DNA sequence. It typically involves the addition of a methyl group to cytosine bases, producing 5-methylcytosine (5-mC).1 In mammalian genomes, DNA methylation is most common at CpG islands—regulatory genomic regions with high numbers of cytosine-guanine dinucleotide repeats.4 Although less common, non-CpG methylation does occur in specific cell types, such as embryonic stem cells, pluripotent stem cells, and neurons.4
DNA methylation mechanisms
A conserved family of cytosine methylase enzymes called DNA methyltransferases (DNMTs) catalyze DNA methylation, typically transferring methyl groups from a molecule known as methyl donor S-adenosine methionine (SAM) to a cytosine’s fifth carbon position.2,3 DNMT3A and DNMT3B can establish de novo methylation on unmodified DNA, while DNMT1 carries out maintenance methylation by transferring the methylation pattern from the parent DNA strand to the daughter strand during DNA replication.1
DNA can also be demethylated, either through passive or active processes. Passive DNA demethylation occurs when the methylation pattern is not replenished during DNA replication and gradually disappears over time.1 In contrast, active DNA demethylation involves enzymatic methyl group removal from cytosine residues. For example, ten eleven translocation (TET) enzymes can oxidize 5-mC to 5-hydroxymethylcytosine (5-hmC).5
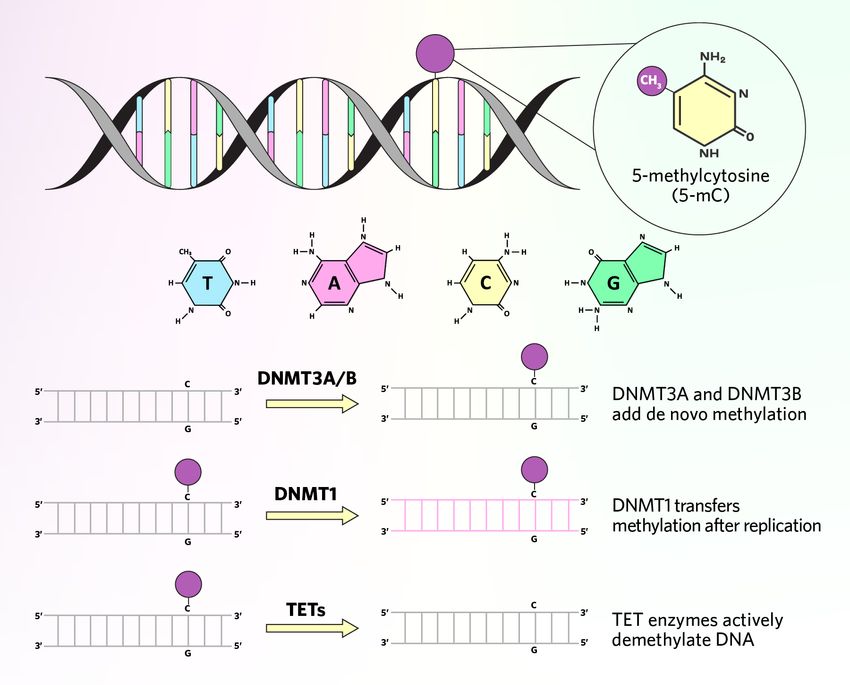
DNA methylation occurs via DNMTs that add methyl groups to cytosine bases, producing 5-mC de novo or transferring 5-mC marks from parental to daughter DNA strands during replication. DNA can become demethylated, either passively if the pattern is not passed down during DNA replication, or actively by DNA demethylating enzymes such as TETs.
Modified from © iStock, olando_o
What Does DNA Methylation Do?
After discovering that DNA methylation frequently occurs in the promoter regions of genes, scientists have attempted to answer a key question: How does DNA methylation affect gene expression? Gene promoter 5-mC DNA methylation represses transcription and causes gene silencing. It does so by affecting the chromatin structure in which DNA is packaged and preventing transcription factors from binding to the affected DNA.1
Along with other epigenetic marks such as histone modification, CpG DNA methylation is responsible for cell- and tissue-specific gene expression. In mammals, DNA methylation is crucial during embryonic development, contributing to cell and tissue differentiation. DNA methylation is also involved in X-chromosome inactivation, silencing of retroviral elements, and genomic imprinting.1 Altered CpG DNA methylation patterns are involved in cancer, autoimmune diseases, neurological disorders, aging, and other conditions. Non-CpG methylation is less studied and its function remains largely unclear.1
DNA Methylation Tests
Scientists can use several different DNA sequencing methods to study DNA methylation and its effects. Bisulfite sequencing, which involves DNA denaturation and treatment with sodium bisulfite, has long been regarded as the gold standard for DNA methylation sequencing and is still widely used.6 However, the chemical reaction damages DNA, leading researchers to develop various bisulfite-free methods for DNA methylation sequencing.6
For example, enzymatic methyl-Seq (EM-Seq) uses several different enzymes to indirectly map 5-mC and 5-hmC modifications. In contrast, TET-assisted pyridine borane sequencing (TAPS) detects 5-mC and 5-hmC modifications directly.6
After sequencing, scientists can perform DNA methylation analysis; by identifying where there are aberrant DNA methylation patterns in the genome, researchers can investigate which genes are affected and how these patterns play a role in biological processes.
How Does DNA Methylation Affect Human Health?
DNA methylation plays a profound role in human development, aging, and health.
DNA methylation in cancer
Perhaps one of the best studied effects of aberrant DNA methylation is in tumorigenesis. Global DNA methylation disruption is a hallmark of cancer; hypomethylation can occur at a genome-wide scale, resulting in genomic instability, while hypermethylation can occur at specific genes.7 Loss of DNA methylation can be correlated with disease severity and the likelihood of metastasis in many cancers.7
DNA methylation in autoimmunity
Alterations to DNA methylation patterns can also contribute to autoimmune disorders. For example, scientists have observed decreased global DNA methylation in the T cells of people with systemic lupus erythematosus (SLE), a complex autoimmune disorder.2 DNMT1 inhibitors cause hypomethylation and subsequent autoreactivity in these immune cells, suggesting a causal relationship between DNA hypomethylation and SLE.
DNA methylation in neurological conditions
Aberrant DNA methylation is also involved in many brain conditions, including neuropsychiatric illnesses, neurodevelopmental disorders, and neurodegenerative diseases.8 In Alzheimer’s disease, scientists have identified both hypo- and hypermethylation in the promoter regions of Alzheimer’s-related genes.8 Additionally, hypomethylation of the alpha-synuclein (SNCA) gene, particularly at intron 1, leads to SNCA overexpression in patients with Parkinson’s disease.8
A more unusual example is Rett syndrome (RTT), a rare neurodevelopmental disorder that involves a period of developmental regression and subsequent motor skill, communication, growth, and cognitive impairments. Scientists discovered that RTT is caused by mutations in methyl CpG binding protein 2 (MeCP2), a DNA methylation reader protein highly expressed in the brain.9 Although mutations in MeCP2 occur at the gene level, they alter the protein’s stability and DNA binding affinity, preventing it from binding to methylated DNA and repressing transcription.10
Final Thoughts on DNA Methylation
DNA methylation is a crucial part of the human epigenome and is responsible for a range of normal biological processes, as well as the onset of many different diseases. As DNA methylation sequencing technologies continue to improve, scientists will discover more DNA methylation functions, human health effects, and potential therapeutic avenues.
FAQ
What does DNA methylation do?
- DNA methylation within gene promoter regions represses transcription and causes gene silencing by affecting chromatin structure and transcription factor binding. CpG DNA methylation helps regulate cell- and tissue-specific gene expression, such as X-chromosome inactivation, silencing of retroviral elements, and genomic imprinting.
Why is DNA methylation important?
- DNA methylation helps dictate how genes are expressed, contributing to normal cellular differentiation during development and aging. Additionally, aberrant DNA methylation is involved in many different disease processes, including cancer, neurological disorders, and autoimmune disease.
What is the difference between DNA methylation and histone methylation?
- DNA methylation and histone methylation are both epigenetic changes that influence gene expression without directly modifying the DNA sequence. DNA methylation involves the addition of methyl groups to the DNA base cytosine, while histone methylation is a posttranslational modification on histone proteins, typically at lysine and arginine residues.
- Moore LD, et al. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23-38.
- Lanata CM, et al. DNA methylation 101: What is important to know about DNA methylation and its role in SLE risk and disease heterogeneity. Lupus Sci Med. 2018;5(1).
- Lyko F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nature Reviews Genetics. 2018;19:81-92.
- Ramasamy D, et al. Non-CpG methylation—A key epigenetic modification in cancer. Brief Funct Genomics. 2021;20(5):304-311.
- Wu X, Zhang Y. TET-mediated active DNA demethylation: Mechanism, function and beyond Nature Reviews Genetics. 2017;18:517-534.
- Chen X, et al. Mapping epigenetic modifications by sequencing technologies. Cell Death Differ. 2025;32(1):56-65.
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597-610.
- Rasmi Y, et al. The role of DNA methylation in progression of neurological disorders and neurodegenerative diseases as well as the prospect of using DNA methylation inhibitors as therapeutic agents for such disorders. IBRO Neurosci Rep. 2022;14:28-37.
- Kriaucionis S, Bird A. DNA methylation and Rett syndrome. Hum Mol Genet. 2003;12(suppl_2):R221-R227.
- Good KV, et al. MeCP2: The genetic driver of Rett syndrome epigenetics. Front Genet. 2021;12.













