Drug development still faces numerous challenges despite the industry’s many scientific advances, resulting in around 90 percent of drugs not making it through clinical trials to approval.1 Many potential novel therapies fail because the target selected is not actually the most promising candidate for the purpose of drug development.2 Growing evidence shows that having supporting data from large-scale human genetics studies for a biological target more than doubles the chances of success.2-4
In particular, genome-wide association studies (GWAS) have pinpointed hundreds of genes associated with disease risk, enabling the development of drugs targeting a broad range of highly prevalent diseases, including rheumatoid arthritis (Abatacept; target: CTLA-4), hypercholesterolemia (Evolocumab; target: PCSK9), and ankylosing spondylitis (Secukinumab; target: IL-17A/IL-23 pathway). Human genetics have also uncovered opportunities for drug repurposing, such as Ustekinumab and Denosumab, originally developed for psoriasis and osteoporosis, which are now successfully used in Crohn’s disease treatment following genetic discoveries.5,6 However, these success stories of human genetics-driven drug discovery have thus far largely been limited to sequence variants in or near protein encoding genes, which comprise only about two percent of the human genome.
Notably, the noncoding part of the human genome—sometimes dubbed the dark genome—harbors around 95 percent of all disease-associated variants uncovered by GWAS,7 suggesting that the noncoding human genome has a vast, yet so far largely untapped, potential to accelerate the discovery of novel therapies to improve population health. Towards this aim, a major challenge has been to translate the statistical hits in these regions from large-scale human genetics studies into functional insights that explain the underlying molecular mechanisms of disease and disease susceptibility.
Now, nearly 25 years after the publication of the first draft of the human genome,8,9 key advances in functional genomics and computational approaches have provided us with a powerful set of tools to fully leverage the huge wealth of human genetics data by making the vast space of the human noncoding genome accessible, and thus actionable for therapy discovery. Approaches harnessing 3D genomics—the high-resolution profiling of the cell-type-specific 3D organization of the human genome—are central to this endeavor, as the technology holds the key to unveiling causal and disease-relevant interactions between the coding and noncoding parts of the human genome.
3D genomics was initially fuelled by the discovery that enhancers, key gene regulatory elements in the noncoding genome that control when and where genes are switched on during human development, can regulate their target genes over large genomic distances, in some cases more than a million base pairs. They do so through a process called chromatin looping, whereby enhancers are brought into close spatial proximity to their target genes, often bypassing more proximal genes.10 This chromatin looping is cell type specific, occurring where these genes need to be expressed. Importantly, a failure to consider this process can lead to the assignment of enhancers to the “wrong” target gene or genes.
A striking example is the human fat mass and obesity-associated (FTO) gene locus, which harbors the strongest genetic association with obesity in the human genome.11,12 As the genetic association was mapped to intronic regions of FTO, the gene itself was initially designated as an obesity susceptibility gene. However, subsequent studies showed that the obesity-associated FTO variants directly interact with the distal IRX3 and IRX5 genes,13 and the predicted causal T-to-C single nucleotide variant (rs1421085) correlates with increased expression levels of IRX3 and IRX5, but not FTO, in human adipose precursors.14 Thus, the obesity-associated region within FTO acts as a functional enhancer for the developmental transcription factor genes IRX3 and IRX5, located approximately 500kb and 1.1Mb away, respectively (below).
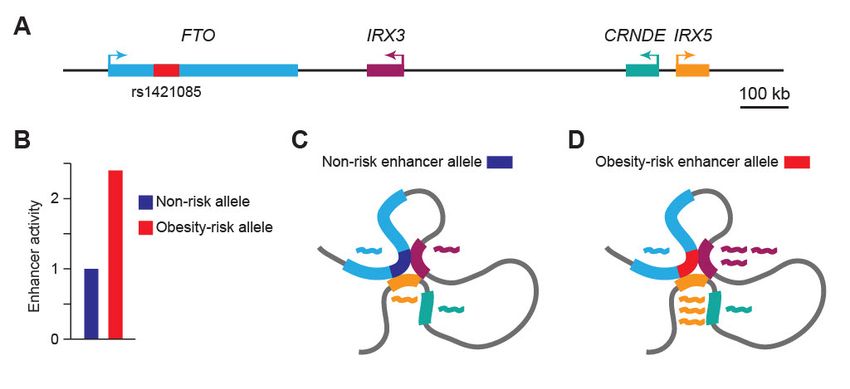
3D genome conformation and gene expression control at the human FTO locus. A) The FTO gene locus encompasses the genes FTO (light blue), IRX3 (burgundy), CRNDE (teal), and IRX5 (orange), with the obesity-risk SNP (rs1421085) containing enhancer located within the FTO gene (red).B) The obesity-risk enhancer allele has stronger enhancer activity than the non-risk allele (modified from reference 14).C) and D) 3D genome folding of the FTO locus (modified from reference 13). The obesity risk enhancer allele (red; D) drives increased expression of IRX3 and IRX5 compared to the non-risk enhancer allele (blue; C). Genes are shown in the same colors as in A), with the corresponding RNA transcripts depicted as wavy lines in the same colors.
The example described above demonstrates the power of combining 3D genomics with orthogonal functional genomics approaches to measure gene expression, chromatin accessibility, and epigenetic marks—an approach we term 3D multiomics—to reveal new disease-associated target genes, which other approaches have been unable to detect. Through a similar approach, a single-nucleotide polymorphism (SNP) in an enhancer associated with multiple immune disorders was recently shown to act on the distant ETS2 gene.15 Enhancers are known to be the primary source of genetic variation associated with human disease,16 so these examples very likely only represent the tip of the iceberg, especially as recent findings suggest that we are still far from reaching peak genetics insights.4
A 3D multiomics approach has the potential to impact a wide range of different disease areas, including autoimmune conditions and diseases that cause dementia. Alzheimer’s Research UK have reported that 55 million people are living with dementia worldwide, and if nothing changes, one in two people will be affected by dementia either as a carer, through developing the disease themselves, or both. Over the past two decades, increasingly powerful GWAS datasets have been reported for conditions such as Alzheimer’s disease, and the data generated have triggered a revolution in dementia drug discovery. As a result of these insights, drug discovery scientists have increased their focus on mechanisms such as the role of microglia and inflammation in the development of neurodegenerative diseases, complementing earlier approaches aimed at neurons and the aggregation and clearance of proteins such as β-amyloid and tau.17
Dementia researchers have used a variety of techniques to link mutations identified in GWAS studies to specific genes, which have subsequently become targets for drug discovery. The application of 3D multiomics to these datasets has the potential to uncover new genes and mechanisms, some of which may operate through cell types other than neurons and microglia.
Intriguingly, enhancers can not only uncover novel genes for drug development through their long-range chromatin contacts, they can also themselves be the target of therapeutic interventions. Casgevy, the first approved genome-editing therapy using CRISPR,18 works by modifying the activity of the blood-cell-specific BCL11A enhancer to increase the production of hemoglobin, offering a curative treatment for patients suffering from beta thalassaemia and sickle cell disease.19
Collectively, these breakthroughs mark a new era of genetics-driven drug discovery, harnessing the noncoding genome to advance personalized medicine. Looking ahead, the integration of 3D multiomics with machine learning has the potential to further accelerate this progress, by predicting enhancer–gene interactions in silico to prioritize targets for validation. While challenges such as data standardization and ethical oversight remain, the convergence of AI and 3D multiomics promises to transform how we discover and develop therapies for complex diseases.
- Kim E, et al. Factors affecting the success of new drug clinical trials. Ther Innov Regul Sci. 2023;57(6):737-750.
- Razuvayevskaya O, et al. Genetic factors associated with reasons for clinical trial stoppage. Nat Genet. 2024;56(12):1862-1867.
- Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47(8):856-860.
- Minikel EV, et al. Refining the impact of genetic evidence on clinical success. Nature. 2024;629(8010):624-629.
- Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118-1125.
- Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979-986.
- Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190-1195.
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860-921.
- Venter JC, et al. The sequence of the human genome. Science. 2001;291(5507):1304-1351.
- Schoenfelder S, Fraser P. Long-range enhancer–promoter contacts in gene expression control. Nat Rev Genet. 2019;20(8):437-455.
- Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889-894.
- Dina C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724-726.
- Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371-374.
- Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373(10):895-907.
- Stankey CT, et al. A disease-associated gene desert directs macrophage inflammation through ETS2. Nature. 2024;630(8018):447-456.
- Robert S, Rada-Iglesias A. The interaction between enhancer variants and environmental factors as an overlooked aetiological paradigm in human complex disease. Bioessays. 2023;45(10):e2300038.
- De Strooper B, Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164(4):603-615.
- Pacesa M, Pelea O, Jinek M. Past, present, and future of CRISPR genome editing technologies. Cell. 2024;187(10):1076-1100.
- Singh A, et al. Revolutionary breakthrough: FDA approves CASGEVY, the first CRISPR/Cas9 gene therapy for sickle cell disease. Ann Med Surg (Lond). 2024;86:4555-4559.
















