Aging comes with loss. Hair becomes thinner, hearing duller, and memory scanter. One of the key regulators of total cellular metabolism, nicotinamide adenine dinucleotide (NAD+), also dwindles.1 “In aging, you may have at most a 30-50 percent decline in NAD+ in different tissues, and we don’t really know which NAD+-dependent processes are likely to be susceptible to that,” said Joseph Baur, a physiologist and expert in NAD+ biology at the University of Pennsylvania Perelman School of Medicine. An electron acceptor essential to many metabolic processes, NAD+ also acts as a co-substrate for enzymes unrelated to redox, implicating NAD+ in DNA repair, signaling, and transcription.1,2
Just as we need to learn to cope with aging, so do our cells. Mathias Ziegler, a medical biochemist at the University of Bergen, and his team are interested in how aging cells deal with NAD+ depletion. According to their research, recently published in Nature Metabolism, it’s an inside job.3

Ziegler’s research group studies molecular signaling and bioenergetics, including NAD+ biology.
Photograph by Torstein Ravnskog; Provided by Mathias Ziegler
NAD+ is compartmentalized into organelles including mitochondria, peroxisomes, and endoplasmic reticulum,4,5 and these partitioned pools regulate various physiological processes.6 Biologically, mitochondrial function and NAD+ levels both decrease with age. “We have generated a model as simple as a cell line to recapitulate these observations,” Ziegler said.
Rather than using pharmacological agents to acutely decrease NAD+ levels, Ziegler and his team developed a chronic NAD+ depletion model by overexpressing the catalytic domain of NAD+-consuming PARP1 (PARPcd) in distinct subcellular compartments in cell lines. This strategy allowed for constitutive NAD+ depletion, similar to what occurs in aging cells, to determine if NAD+ pools in different compartments are connected. They found that PARPcd-expressing cells handled NAD+ depletion well unless the mitochondrial NAD+ pool was targeted. When PARPcd was expressed in mitochondria, the researchers found defects in mitochondrial metabolism.
Given the sensitivity of the mitochondrial NAD+ pool, the researchers examined this compartment more closely. With the expression of PARPcd, mitochondrial NAD+ levels were lower than their wild-type counterparts, even when decreases in NAD+ originated in different compartments. This suggested that mitochondrial NAD+ was siphoned out to other organelles to cope with NAD+ depletion elsewhere. “We have segregated pools, and we don’t really know how they come about…We do know that they interact and that they steal from each other in a way,” Ziegler explained.
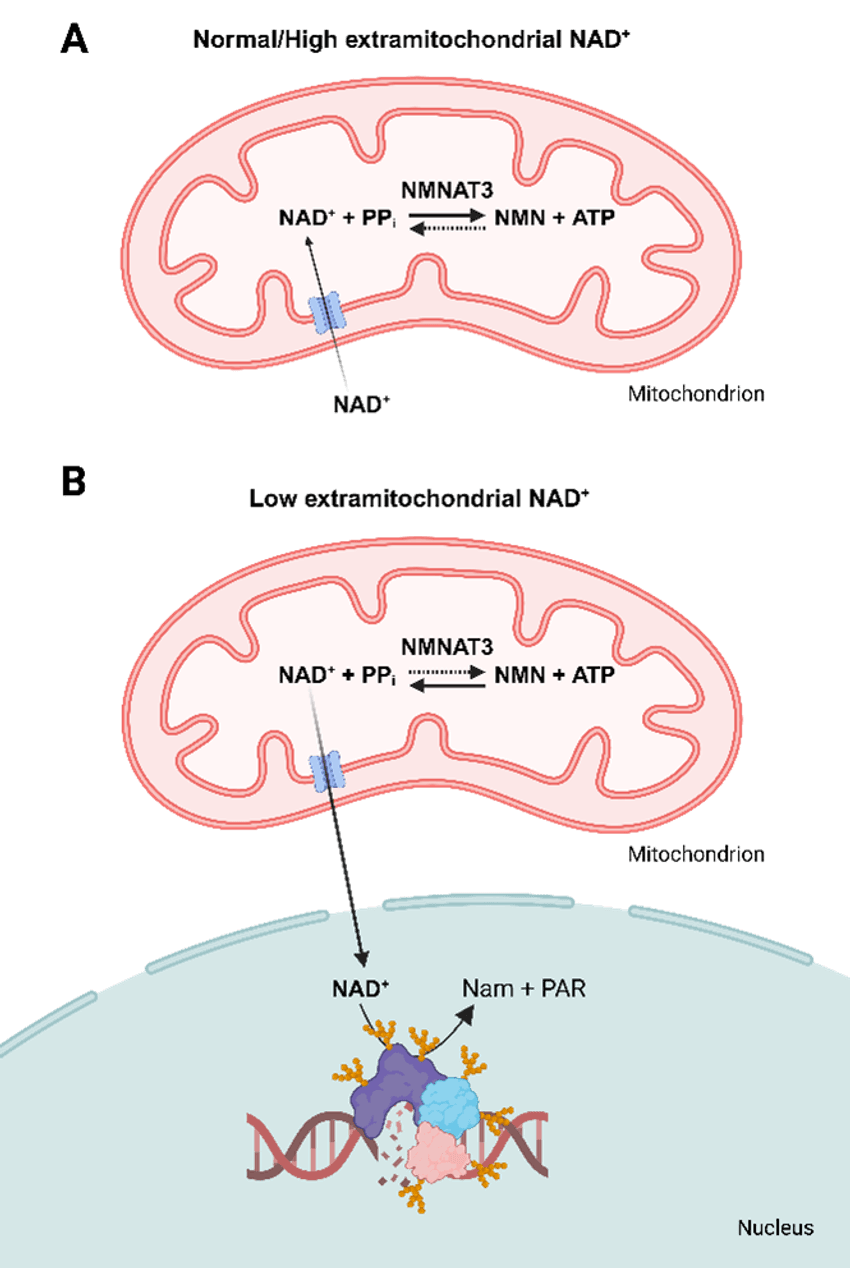
Ziegler’s research group proposed a model to explain how mitochondria alleviate decreases in NAD+ levels: When there are high NAD+ levels in the cell (A), mitochondria import NAD+ and NMNAT3 generates NAD+ equivalents to create a large pool. When there are low NAD+ levels in the cell (B), mitochondria export NAD+ elsewhere, such as the nucleus.
Scheme created and provided by lena Høyland
Considering the role of mitochondria as an NAD+ repository, the team investigated the maintenance of the large mitochondrial NAD+ pool.4 Mitochondria house an isoform of nicotinamide mononucleotide (NMN) adenylyltransferase, NMNAT3, an NAD+ biosynthesis enzyme. However, the mitochondrial NAD+ pool is generated by a recently identified mitochondrial NAD+ transporter,7 not NMNAT3,8 so NMNAT3’s function has remained elusive. “Everyone was scratching their heads a little bit about why do we have this then,” Baur, a previous collaborator of Ziegler’s who was not involved in this study, said regarding NMNAT3.
Ziegler’s group found that NMNAT3 maintains a reservoir of NMN, an NAD+ equivalent, inside mitochondria. Combined with transport of NAD+ into mitochondria, this compartment is poised with a high concentration of NAD+ and NAD+ equivalents to defend against cellular depletion of the cofactor. Baur, who was not involved in this study, was surprised by this result. “That’s I think the first plausible explanation for why we would retain NMNAT3 if it’s not actually used to generate the NAD+ pool in the first place,” Baur said.
The tools generated by Ziegler and his team establish mitochondria as a rheostat, or tuner, for cellular NAD+ levels, to buffer and compensate for cellular NAD+ loss. However, this organelle isn’t a cure-all in the search for the fountain of youth. Alterations in NAD+ homeostasis can affect age-related pathologies such as neurodegeneration and cancer,1,2 but the mechanisms and implications of this remain unclear.9 Mitochondria represent a potential vulnerability that could be contributing to age-related diseases, motivating further study of compartmentalized NAD+ pools and NAD+ transport. For now, while some might try to delay the inevitable wrinkles and gray hair, people can rest assured that the mitochondria are battling against cellular NAD+ depletion in aging cells.
- Katsyuba E, et al. NAD+ homeostasis in health and disease. Nat Metab. 2020;2(1):9-31.
- Cantó C, et al. NAD+ metabolism and the control of energy homeostasis: A balancing act between mitochondria and the nucleus. Cell Metab. 2015;22(1):31-53.
- Høyland LE, et al. Subcellular NAD+ pools are interconnected and buffered by mitochondrial NAD+. Nat Metab. 2024;6(12):2319-2337.
- Cambronne XA, et al. Biosensor reveals multiple sources for mitochondrial NAD+. Science. 2016;352(6292):1474-1477.
- Dölle C, et al. Visualization of subcellular NAD pools and intra-organellar protein localization by poly-ADP-ribose formation. Cell Mol Life Sci. 2010;67(3):433-443.
- Cambronne XA, Kraus WL. Location, location, location: Compartmentalization of NAD+ synthesis and functions in mammalian cells. Trends Biochem Sci. 2020;45(10):858-873.
- Luongo T, et al. SLC25A51 is a mammalian mitochondrial NAD+ transporter. Nature. 2020;588(7836):174-179.
- Yamamoto M, et al. Nmnat3 Is Dispensable in Mitochondrial NAD Level Maintenance In Vivo. PLoS ONE. 2016;11(1):e0147037.
- McReynolds MR, et al. Age-related NAD+ decline. Exp Gerontol. 2020;134:110888.