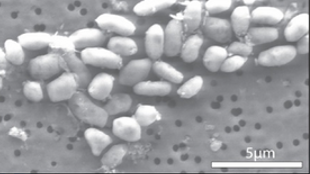
 The newly-sequenced GFAJ-1
The newly-sequenced GFAJ-1
The GFAJ-1 bacterium, which whipped up controversy a year ago when researchers claimed that it could survive and grow using arsenic instead of phosphorus, has been sequenced. Scientists from the University of Illinois, Chicago, (UIC) and elsewhere posted GFAJ-1's genome sequence—which contains 3,400 genes within its 3.5 million basepairs—in Genbank last week.
UIC arsenic microbiologist Simon Silver, a vocal critic of the claim that the bacteria can thrive on arsenic, told ScienceInsider that the sequence doesn't settle the debate over whether the organism can incorporate arsenic into its DNA, but it does reveal that GFAJ-1 has fewer genes known to help organisms survive in high-arsenic environments than E. coli does. Silver added that he's not hopeful that the GFAJ-1 sequence will convince the researchers who ...




















