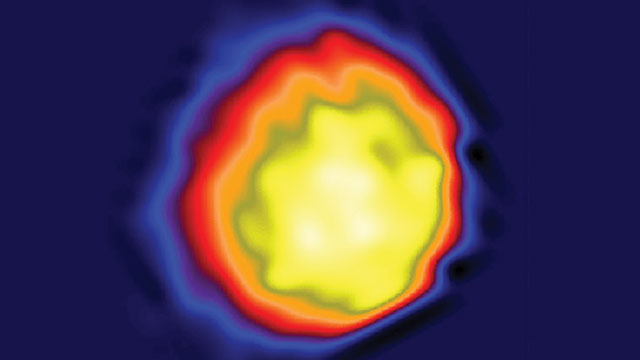
 NAKED VIRUS: The first high-contrast image of a single herpesvirus ever taken with a coherent X-ray synchrotron. The virus, with its capsid, measures about 100 nm in diameter.COURTESY OF JIANWEI MIAO
NAKED VIRUS: The first high-contrast image of a single herpesvirus ever taken with a coherent X-ray synchrotron. The virus, with its capsid, measures about 100 nm in diameter.COURTESY OF JIANWEI MIAO
It was just over a century ago that William Lawrence Bragg and his father, William Henry Bragg, kick-started the science of X-ray crystallography in a talk at the Cambridge Philosophical Society on November 11, 1912. Since then, thousands upon thousands of structures have been solved, from table salt and diamond to RNA polymerase and the ribosome.
Back in the Braggs’ time, crystals were analyzed using laboratory X-ray tubes, which are relatively weak sources of continuous, noncoherent light—that is, light that travels in all directions, like lamplight. In the 1970s, researchers started using synchrotron particle accelerators, which can shoot partially focused (i.e., relatively coherent), highly intense X-ray radiation at a crystal. According to Jianwei Miao, a professor of physics and astronomy at the University of California, ...




















