Bacteria are among the most diverse lifeforms on Earth, so it’s no surprise that their genomes have yielded a treasure trove of fascinating discoveries. The study of bacterial genomes has led to the discovery of bacterial multicellular immunity, an understanding of unusual forms of metabolism, and even the possibility of tweaking the human microbiome for improved health outcomes. In recent years, researchers have been hard at work developing targeted tools to sequence and genetically modify bacterial genomes, contributing to a burgeoning field of research.
A Hidden World of Bacterial Genomes in Our Food
Using shotgun metagenomics, researchers have compiled the largest database of food-associated microbes to date, enabling the identification of diverse microbial species present in certain foods. Compiled from samples of over 2,500 foods from 50 different countries, the database includes more than 1,100 bacterial genomes. Almost half of these microbes live exclusively on specific foods and are not found in any other environment, while a similar number are entirely new to science. Because some of the bacterial genomes identified are associated with food spoilage, the study provides a basis for quality control and food spoilage tests, a better understanding of some strains found in the human microbiome and could potentially be applied in waste reduction strategies to extend the shelf-life of foods.
Explore the diversity of bacterial genomes in our food here.
Studying Rare Bacterial Genomes with a New Sequencing Tool

Gang Fang’s team developed mEnrich-seq, a sequencing technique that enriches bacteria of interest in a sample.
Brian Schutza
Identifying and sequencing rare strains of bacteria that make up the human microbiome has been challenging for scientists. Gang Fang, a geneticist at the Icahn School of Medicine at Mount Sinai who has based his career on the sequencing of bacterial genomes, developed a technique called mEnrich-seq to address this problem. By utilizing the unique epigenetic signatures of specific bacterial strains, mEnrich-seq can boost the signal of strains of interest by over 100-fold, preventing them from being drowned out by the ‘noise’ of abundant species. This method of bacterial genome sequencing could be used to identify antibiotic-resistant strains—for example, in patients with urinary tract infections—or to locate beneficial strains in fecal samples that could be used as probiotics.
Read more about the mEnrich-seq method in this article.
Engineering Bacterial Genomes to Improve the Human Microbiome
The human microbiome has a range of effects on our health and well-being, prompting researchers to investigate how modulating the genomes of these microbes can improve health outcomes. The advent of CRISPR gene editing technology has made it possible for scientists to modify a range of bacterial genomes, but not all species are amenable to editing. A team of bioengineers from the University of California, Berkeley developed a sequencing method to identify genetically malleable bacterial genomes, then tinkered with CRISPR technology to create an all-in-one DNA editing system they could apply to those species. Using this approach, they were able to enrich specific strains within stool samples, highlighting its potential for therapeutic applications.
Delve into the methods the team developed for editing bacterial genomes in this story.
A Giant Bacterial Genome Contains Genes for an Unusual Metabolism
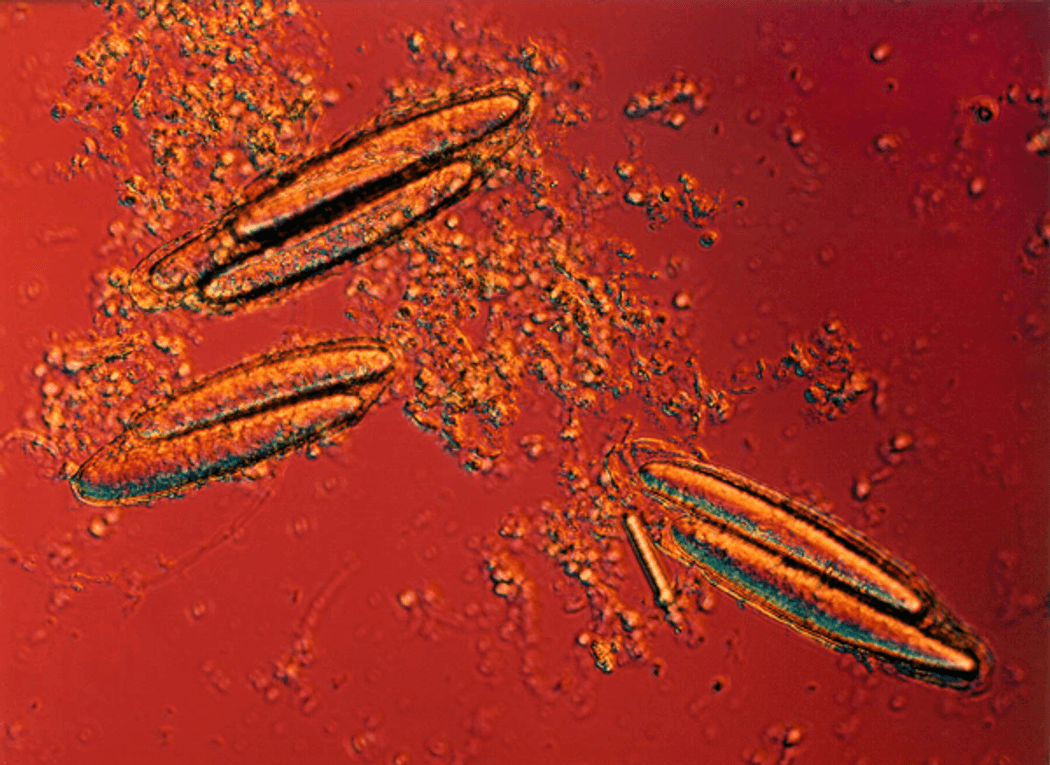
Insights into the genomic material of the giant bacterium shed light on how this organism gathers energy.
Ester Angert
Once mistaken for a protist, the giant bacterium Epulopiscium viviparus recently had its genome sequenced for the first time, revealing some unusual traits that allow it to gather energy. Using genomics and transcriptomics, microbiologist David Sannino and his team at the University of Glasgow discovered that E. viviparous lacks genes that are specific for respiration, the metabolic mechanism that yields the most energy. Instead, the giant bacterium powers its metabolism using the combination of several less efficient metabolic processes, such as fermentation and sodium-motive force.
Explore more of the team's discoveries about the giant bacterial genome in this article.
Multicellular Bacterial Genomes Reveal Complex Mechanisms for Immunity
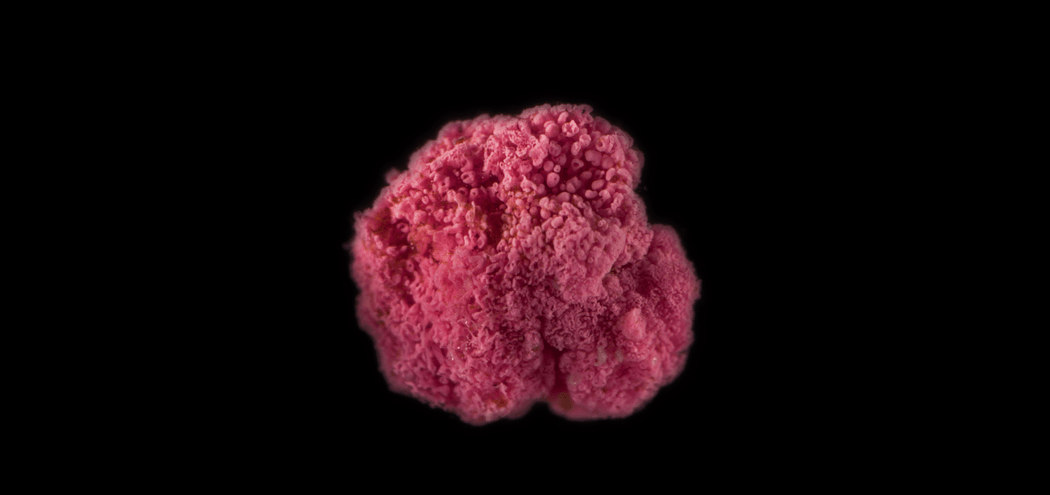
Researchers explored how purple sulfur bacteria that clump together to form pink berries protect themselves from infectious agents.
Scott Chimileski, Marine Biological Laboratory
A study of multicellular bacterial genomes demonstrated how a rare mutagenesis system known as diversity-generating retroelements (DGR) allows cooperating groups of microbes to defend themselves from infectious agents. Structurally similar to antibodies produced by the human immune system, DGR proteins recognize invaders like bacteriophages and can hypermutate specific sites in the bacterial genome. Microbiologist Elizabeth Wilbanks, and her colleagues at the University of California, Santa Barbara sequenced the genomes of bacteria found in aggregates called pink berries to explore their DGR repertoire. The DGR found in these bacteria makes them capable of producing more variations in target genes than there are protons in the universe, but how they protect the bacteria from pathogens is yet to be explored.
Discover more about DGR systems in multicellular bacterial genomes here.
As better tools are developed and new strains are sequenced, researchers will continue to make discoveries about bacterial genomes that contribute to our understanding of microbial communities, the environment, and human health. With gene editing tools like CRISPR, bioengineers may be able to modify the human microbiome and develop targeted therapeutics.













