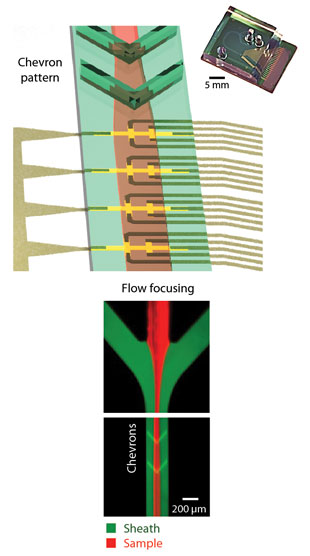
 IT’S MAGNETIC: Hakho Lee’s CTC counter features eight micro-Hall elements (yellow crosses, above), staggered to maximize detection accuracy. Flowing “sheath fluid” focuses the cells (right), while architectural elements in the channel (chevrons) force them towards the bottom of the channel. A photo of the physical chip is shown at top right.D. ISSADORE ET AL., SCIENCE TRANSL MED, 4:141ra92, 2012. Originally an immunology tool and a fixture of cell biology research for decades, flow cytometry allows users to catalog a dozen or more molecular and physical features in a single cell. A related, more sophisticated instrument, the fluorescence-activated cell sorter (FACS), enables researchers to isolate particular cells from heterogeneous populations.
IT’S MAGNETIC: Hakho Lee’s CTC counter features eight micro-Hall elements (yellow crosses, above), staggered to maximize detection accuracy. Flowing “sheath fluid” focuses the cells (right), while architectural elements in the channel (chevrons) force them towards the bottom of the channel. A photo of the physical chip is shown at top right.D. ISSADORE ET AL., SCIENCE TRANSL MED, 4:141ra92, 2012. Originally an immunology tool and a fixture of cell biology research for decades, flow cytometry allows users to catalog a dozen or more molecular and physical features in a single cell. A related, more sophisticated instrument, the fluorescence-activated cell sorter (FACS), enables researchers to isolate particular cells from heterogeneous populations.
These instruments have had a profound impact on biological research. Yet there is much they cannot do. For instance, with some exceptions, they can only count or sort cells based on their protein content. Thus, researchers cannot easily use the technique to identify cells that contain, say, specific mutations. Researchers also have to know in advance what molecules they are looking for, and have fluorescently labeled antibodies available to target them. And because the method depends on antibody binding to surface receptors, there’s always the possibility that sorted cell populations will be activated or altered by the process itself.
“For the most part, flow cytometry is used when you have a good affinity assay—an antibody or oligomer probe that can specifically label your cell type,” ...




















