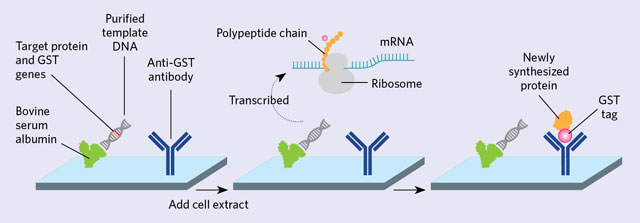
 BUILT ON THE SPOT: Purified template DNA is printed onto a slide along with bovine serum albumin and an antibody that recognizes glutathione-S-transferase (GST). The DNA template contains both the gene for the protein of interest and for GST (left). LaBaer isn’t sure why bovine serum albumin helps, but it seems to hold the DNA to the surface without inhibiting transcription and may provide some degree of background suppression. Transcription and translation is initiated by the addition of a cell extract, and the protein self-assembles (middle). The GST-tagged protein is then captured by an anti-GST antibody (right).ADAPTED FROM WIKIPEDIA/SIMIN LIM
BUILT ON THE SPOT: Purified template DNA is printed onto a slide along with bovine serum albumin and an antibody that recognizes glutathione-S-transferase (GST). The DNA template contains both the gene for the protein of interest and for GST (left). LaBaer isn’t sure why bovine serum albumin helps, but it seems to hold the DNA to the surface without inhibiting transcription and may provide some degree of background suppression. Transcription and translation is initiated by the addition of a cell extract, and the protein self-assembles (middle). The GST-tagged protein is then captured by an anti-GST antibody (right).ADAPTED FROM WIKIPEDIA/SIMIN LIM
Thirty years after it came into use, the protein microarray—a high-throughput tool that tracks the presence, activity, and interactions of proteins—is still going strong, even as its DNA counterpart is being replaced by next-generation sequencing. Protein arrays are workhorses in biomarker discovery, immune-response monitoring, and other areas of biology.
Traditional protein microarrays—slides, beads in a liquid, or plates that are specked with hundreds or thousands of proteins, probed and analyzed, often with fluorescent labels—allow an unbiased look at protein function. Unlike other proteomics techniques, arrays immobilize even low-abundance proteins. “The nice thing about protein arrays is that every protein gets its chance. Every protein is there at reasonable levels,” says Josh LaBaer, director of the Virginia G. Piper Center for Personalized Diagnostics at Arizona State ...
















