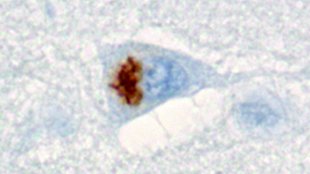
 Dipeptide repeat inclusion in patient brainKohji Mori and Dieter EdbauerA repetitive DNA sequence that was not believed to encode proteins is, in fact, the source of insoluble peptide chains that aggregate in the brain cells of patients displaying certain types of neurodegeneration, according to a study published today (February 7) in Science. These aggregates occur in a wide range of neurodegenerative disorders, so determining their identity is an important first step towards understanding how they might contribute to various pathologies.
Dipeptide repeat inclusion in patient brainKohji Mori and Dieter EdbauerA repetitive DNA sequence that was not believed to encode proteins is, in fact, the source of insoluble peptide chains that aggregate in the brain cells of patients displaying certain types of neurodegeneration, according to a study published today (February 7) in Science. These aggregates occur in a wide range of neurodegenerative disorders, so determining their identity is an important first step towards understanding how they might contribute to various pathologies.
“It’s a very exciting and important paper,” said Bruce Miller, a professor of neurology at the University of California, San Francisco, who was not involved in the study. “We’ve all been waiting, in the field, for someone to make this breakthrough, so I’m just thrilled.”
FTLD-ALS spectrum disorders are a range of related neurodegenerative disorders from frontotemporal lobar degeneration (FTLD) right through to amyotrophic lateral sclerosis (ALS). Most cases are of unknown origin, but an expanded repeat region in a non-coding part of a gene called C9orf72 “is the most prevalent cause we know of for both FTLD and ALS,” Miller said. Almost all patients with FTLD-ALS have characteristic protein aggregates, or inclusions, in their brain cells, but patients with the C9orf72 mutation have an ...



















