Located at the cellular interface, membrane proteins play critical regulatory roles in the signaling between a cell and its interacting environment, making them popular and ideal drug targets. Although methodological limitations have been a bottleneck for drug development, new research tools are opening the door to the next generation of membrane protein-targeted therapeutics.

Yvonne Tan, PhD
Associate Director of Product
Nuclera
In this Innovation Spotlight, Yvonne Tan, Nuclera’s associate director of product, discusses how the eProtein Discovery™ system’s microfluidic-enabled protein synthesis and lipid nanodisc technology help researchers move beyond technical constraints of conventional membrane protein analysis, from fundamental research to drug discovery.
What is the significance of membrane proteins in drug discovery and development research?
Membrane proteins are the gatekeepers of cellular communication. Embedded in the lipid bilayers of cells, these proteins regulate signaling, transport, and environmental sensing, making them vital to nearly every physiological process. Unsurprisingly, they are also one of the most important classes of drug targets, with over 60 percent of approved pharmaceuticals acting on membrane proteins.1 Despite their importance, these proteins have remained one of the most elusive and difficult classes of biomolecules to study.
In drug discovery, the goal is often to modulate specific molecular functions, many of which are governed by membrane proteins such as G protein-coupled receptors (GPCRs), ion channels, and transporters. These proteins play central roles in diseases ranging from cancer to neurological disorders. They’re incredibly rich with therapeutic potential yet represent one of the most under-explored areas in functional and structural biology.
The reason is simple: while membrane proteins are everywhere, they’re exceptionally hard to work with.
Which methods have researchers historically relied on to study membrane proteins?
Researchers have historically relied on labor-intensive and indirect techniques to study membrane proteins, with limited success. Studying these proteins has traditionally required their extraction from cellular membranes using detergents, followed by time-consuming purification steps. These methods—often reliant on cell culture or expression systems like E. coli, insect, or Chinese hamster ovary (CHO) cells—are not only slow but also prone to failure. An issue with using cell-based systems for expression of membrane proteins is toxicity. Some proteins are naturally toxic to these cellular systems. Toxicity can also arise from the cellular machinery becoming overburdened by expressing high levels of the target protein.
Why are membrane proteins so difficult to investigate?
Even when successfully expressed and isolated, membrane proteins tend to be unstable outside their native lipid environments and can form insoluble aggregates when expressed in the aqueous cytoplasm of cells. As such it is difficult to purify membrane proteins in quantities and conformations suitable for downstream assays such as structural analysis, ligand screening, or functional characterization. This bottleneck has slowed the pace of innovation in drug discovery.
What is Nuclera’s new membrane protein workflow and how does it work?
We have developed a microfluidic-based solution to tackle challenges associated with production of membrane proteins head-on. Our latest advancement—an expansion of the eProtein Discovery™ system—introduces a dedicated membrane protein workflow that enables researchers to study these complex biomolecules rapidly, reliably, and without reliance on traditional cell-based expression systems.
Our approach integrates cell-free protein synthesis with microfluidic technology and lipid nanodiscs. In essence, the workflow co-translationally incorporates newly synthesized membrane proteins into pre-assembled nanodiscs—tiny, discoidal lipid bilayers stabilized by scaffold proteins. These nanodiscs mimic the protein’s native environment, preserving proper folding and functionality.
Active membrane proteins are synthesized within 48 hours, directly usable for binding assays, structural studies, or downstream applications, without requiring detergent purification or reconstitution.
How have scientists used eProtein Discovery™ to study membrane protein biology?
The power of our system is already being demonstrated in cutting-edge research. For instance, Nuclera scientists using eProtein Discovery™ recently explored the expression and functional analysis of the human β2-adrenergic receptor, a prototypical GPCR and a critical target for cardiovascular and respiratory diseases. With our system, they were able to generate receptor-loaded nanodiscs rapidly and evaluate ligand binding in the same experiment—providing real-time insights into receptor-ligand interactions.
Another case involves Nuclera researchers investigating multidrug resistance proteins—key players in chemotherapy resistance. These membrane-bound transporters were expressed and functionally validated using our system , helping identify small molecules that modulate efflux activity. Such findings could pave the way for new strategies in overcoming drug resistance in cancer therapy.
Both examples illustrate how our membrane protein workflow enables high-throughput, hypothesis-driven experimentation that was previously out of reach.
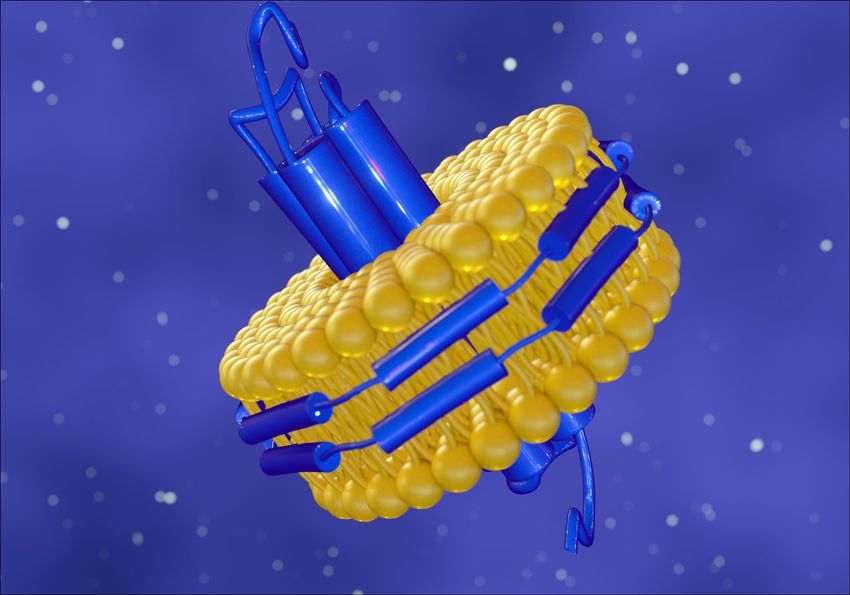
The eProtein Discovery™ system co-translationally incorporates newly synthesized membrane proteins into tiny, discoidal lipid bilayers stabilized by scaffold proteins, mimicking the native environment and preserving membrane protein folding and functionality.
iStock, Love Employee
What excites you the most about the future of microfluidic-based membrane protein workflows?
What excites us most about the future is the scalability and accessibility that microfluidic-based systems like eProtein Discovery™ offer. Scientists no longer need to spend weeks troubleshooting expression systems or purifying proteins. Instead, they can shift focus to experimentation, iteration, and discovery.
Microfluidic technology is democratizing access to high-quality protein studies. By removing technical barriers and compressing timelines, these systems are empowering more labs—from academia to biotech—to interrogate membrane protein biology with unprecedented speed and precision.
Moreover, the flexibility of these systems makes them ideal for difficult-to-express proteins, rare variants, or structure-function screens that were previously cost- or labor-prohibitive.
- Overington JP, et al. How many drug targets are there? Nat Rev Drug Discov. 2006;5(12):993-6.