 THE TECHNIQUE: A gene of interest is hybridized with a biotin-tagged DNA probe (red). Next, an anti-biotin antibody (pink) and an antibody (blue) recognizing an epigenetic mark—e.g., histone H3 methylation—are applied. These antibodies are then each tagged with PLA antibodies (orange and yellow). If the biotin and epigenetic mark are in close proximity, the two PLA antibodies will interact and create a signal detectable with a fluorescent DNA probe.GEORGE RETSECK
THE TECHNIQUE: A gene of interest is hybridized with a biotin-tagged DNA probe (red). Next, an anti-biotin antibody (pink) and an antibody (blue) recognizing an epigenetic mark—e.g., histone H3 methylation—are applied. These antibodies are then each tagged with PLA antibodies (orange and yellow). If the biotin and epigenetic mark are in close proximity, the two PLA antibodies will interact and create a signal detectable with a fluorescent DNA probe.GEORGE RETSECK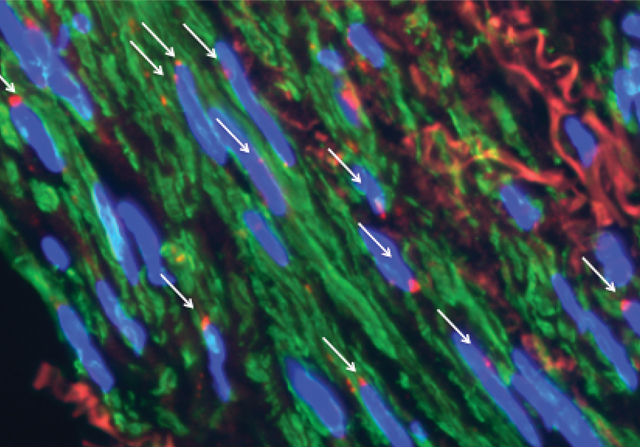 THE PICTURE: Red fluorescent signals (white arrows) appear only in nuclei (blue) where the biotinylated gene probe and epigenetic mark are in close proximity.IMAGE COURTESY OF DELPHINE GOMEZ
THE PICTURE: Red fluorescent signals (white arrows) appear only in nuclei (blue) where the biotinylated gene probe and epigenetic mark are in close proximity.IMAGE COURTESY OF DELPHINE GOMEZ
Techniques exist to visualize specific gene loci within tissue sections. And separate test-tube experiments exist to determine those genes’ epigenetic modifications. Now Gary Owens, a professor of cardiovascular research at the University of Virginia, has devised a new technique that enables gene visualization and epigenetic analysis at the same time.
“The dirty little secret of epigenetics research is that we report quantitative differences from a cell population,” says Andrew Feinberg, a professor of molecular medicine at Johns Hopkins University who was not involved in the study. “If you really want to understand mechanisms, you also need to measure individual cells.”
To achieve single-cell precision, Owens modified an existing technique called a proximity ligation assay (PLA) that is used to determine if two proteins are in ...





















