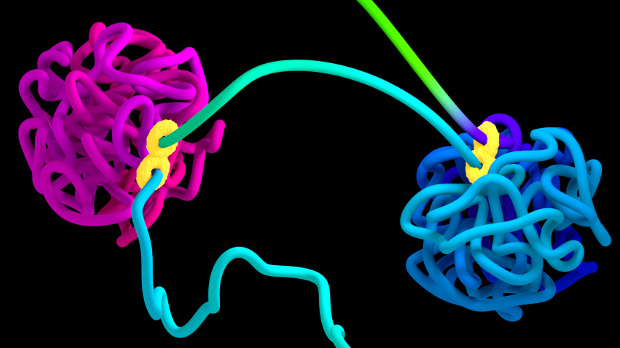
 Two chromatin loops (magenta and blue) held together by a CTCF-containing protein complex (yellow)ADRIAN SANBORN, NAJEEB TARAZI, EREZ LIEBERMAN AIDEN, BAYLOR COLLEGE OF MEDICINE
Two chromatin loops (magenta and blue) held together by a CTCF-containing protein complex (yellow)ADRIAN SANBORN, NAJEEB TARAZI, EREZ LIEBERMAN AIDEN, BAYLOR COLLEGE OF MEDICINE
Compared to its sequence, relatively little is known about the structure of the human genome, which enables more than two meters of chromatin to fit inside the nucleus. Chromosomes are thought to be organized into loops that bring together distant DNA elements—genes, promoters, and enhancers. These chromatin loops are thought to help facilitate gene regulation.
Now, researchers at the Baylor College of Medicine’s Center for Genome Architecture in Houston, Texas, have developed a mathematical model using the binding pattern of a single protein to DNA to predict the looping organization of the human genome. The model helped the team predict the results of its CRISPR-based experimental modification of some of these protein-binding sites in human cells. The team’s results, published this week (October 19) in ...




















