 WIKIMEDIA, RHODODENDRONBUSCHThe CRISPR/Cas9 system of genome editing is exceptionally useful for finding precise sites in the genome to chop up for removing or rewriting genes. But as it stands, the guide RNAs used to home in on the right spots in the genome limit the target sequence to those starting with guanine. Vinod Ranganathan, a postdoc in Donald Zack’s lab at the Johns Hopkins University School of Medicine, and his colleagues reported today (August 8) in Nature Communications that they’ve modified the expression of guide RNAs to also recognize genomic sites starting with adenine.
WIKIMEDIA, RHODODENDRONBUSCHThe CRISPR/Cas9 system of genome editing is exceptionally useful for finding precise sites in the genome to chop up for removing or rewriting genes. But as it stands, the guide RNAs used to home in on the right spots in the genome limit the target sequence to those starting with guanine. Vinod Ranganathan, a postdoc in Donald Zack’s lab at the Johns Hopkins University School of Medicine, and his colleagues reported today (August 8) in Nature Communications that they’ve modified the expression of guide RNAs to also recognize genomic sites starting with adenine.
“Our results enhance the versatility of the CRISPR technology by more than doublingthe number of targetable sites within the human genome and other eukaryotic species,” the team wrote in its paper.
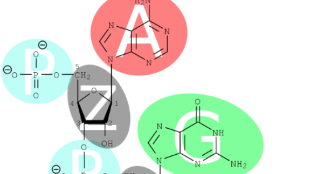
By complementary base pairing, guide RNAs recognize a sequence starting with guanine, continuing with 20 nucleotides, then finishing up with two guanines (GN19NGG). Ranganathan’s new protocol employed a different promoter to express the guide RNA, one that could deliver a transcript with either a guanine or adenine in the first position, meaning that CRISPR can now also break into sites with the sequence AN19NGG. Such sites “occur ~15% more frequently than GN19NGG sites in the human genome and the increase in targeting space is also enriched at human genes and disease loci,” the team ...