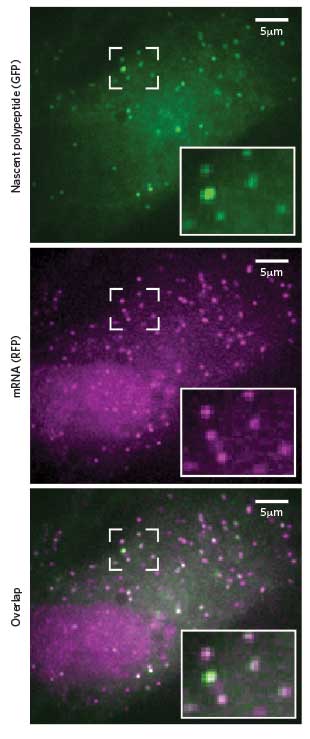
 NEWBORN PROTEINS: Researchers can spot translation in progress by the combined presence of fluorescent tags that stick to newly formed peptides (green) and their corresponding mRNA (pink).Cell, 165:990-1001, 2016Sixty years ago, Francis Crick proposed the central dogma of cell biology: DNA makes RNA makes protein. Back then, watching that sequential process in actual living cells no doubt seemed the stuff of science fiction.
NEWBORN PROTEINS: Researchers can spot translation in progress by the combined presence of fluorescent tags that stick to newly formed peptides (green) and their corresponding mRNA (pink).Cell, 165:990-1001, 2016Sixty years ago, Francis Crick proposed the central dogma of cell biology: DNA makes RNA makes protein. Back then, watching that sequential process in actual living cells no doubt seemed the stuff of science fiction.
But by 1996, researchers had figured out a way to view individual DNA loci in live cells. They engineered the cells’ genomes to contain tandem repeats of a specific nucleotide sequence, introduced green fluorescent proteins that would bind to the repeats, and observed the resulting cluster of fluorescent proteins—which appeared as a bright spot in the nucleus—under the microscope.
Shortly afterwards, scientists came up with a similar trick for visualizing single mRNA molecules: fluorescent antibodies were targeted to a string of repeated stem-loops, RNA sequences designed to fold back on themselves. And by 2004, a few months before Crick’s death, it was possible to combine these two techniques and watch transcription in real time.
“You had an RNA being born from a particular locus of ...



















