Since scientists began directing the evolution of proteins to obtain various desirable outcomes, the tools and techniques used to accomplish these goals have themselves evolved. One recent development, for example, is phage-assisted continuous evolution (PACE), which can generate desired protein variants in a fraction of the time it takes using traditional stepwise evolution methods.
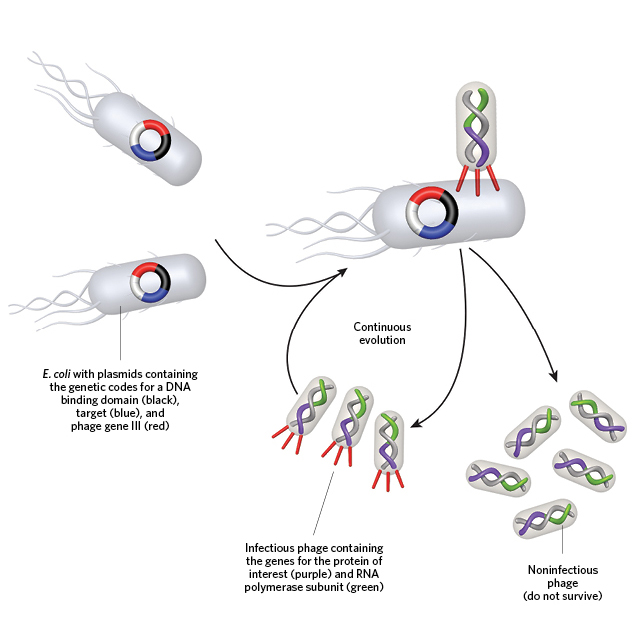 A PERFECT MATCH: To evolve a strong binding affinity between a protein of interest (POI) and a desired target, the gene for the POI (fused to an RNA polymerase subunit) is first encoded into the genome of a bacteriophage lacking a gene (gene III) critical for robust infection of bacteria. These POI-containing viruses are then cultured with E. coli that contain gene III as well as the POI’s desired target (above).© GEORGE RETSECK
A PERFECT MATCH: To evolve a strong binding affinity between a protein of interest (POI) and a desired target, the gene for the POI (fused to an RNA polymerase subunit) is first encoded into the genome of a bacteriophage lacking a gene (gene III) critical for robust infection of bacteria. These POI-containing viruses are then cultured with E. coli that contain gene III as well as the POI’s desired target (above).© GEORGE RETSECK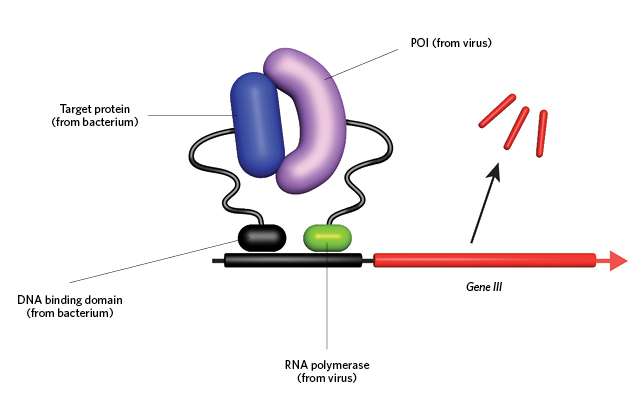 Interaction between the POI and target results in recruitment of the E. coli RNA polymerase to the gene III promoter (black), which drives transcription (above). Thus, only those viruses whose POI evolves a strong binding affinity for the target will be able to drive gene III expression, continuously infect the E. coli, and survive.© GEORGE RETSECK
Interaction between the POI and target results in recruitment of the E. coli RNA polymerase to the gene III promoter (black), which drives transcription (above). Thus, only those viruses whose POI evolves a strong binding affinity for the target will be able to drive gene III expression, continuously infect the E. coli, and survive.© GEORGE RETSECK
And now PACE, too, has evolved. Early examples of PACE were largely used to evolve DNA-binding proteins, which was all well and good, says Greg Weiss of the University of California, Irvine, but the latest incarnation of the technique—protein-binding PACE—is “easily the coolest demonstration of PACE to date.” The ability to evolve novel protein-protein interactions, Weiss explains, “brings the technology into the realm of . . . the therapeutics industry, diagnostics ...





















