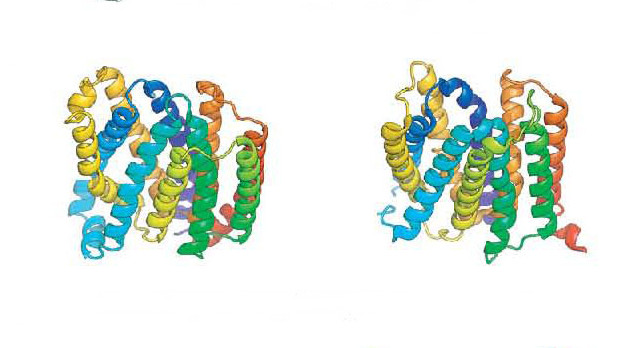
 S. OCHINNIKOV ET AL, SCIENCE 2017DNA sequence data collected from assorted environments has helped researchers generate 3-D models of more than 600 protein families for which the structures were previously unknown, according to a paper published in Science today (January 19). The metagenomic data enabled protein sequence comparisons across an array of species, which lent a statistical power to the predictions that would otherwise not have been possible.
S. OCHINNIKOV ET AL, SCIENCE 2017DNA sequence data collected from assorted environments has helped researchers generate 3-D models of more than 600 protein families for which the structures were previously unknown, according to a paper published in Science today (January 19). The metagenomic data enabled protein sequence comparisons across an array of species, which lent a statistical power to the predictions that would otherwise not have been possible.
“The big take-home message is that it is now possible to use computational methods to get very good models of protein structures,” said protein biochemist David Eisenberg of the University of California, Los Angeles, who was not involved in the study. “That’s a big deal because [the authors] were able to get models for many more proteins than was possible even a few years ago”
Importantly, added computational biologist Johannes Söding of the Max Planck Institute for Biophysical Chemistry in Munich, Germany, who also did not participate in the research, the “method does not need any experimental data,” such as that obtained by X-ray crystallography or nuclear magnetic resonance imaging—classical techniques for revealing a protein’s structure.
Until recently, Söding explained, biologists would predict the structures ...


















