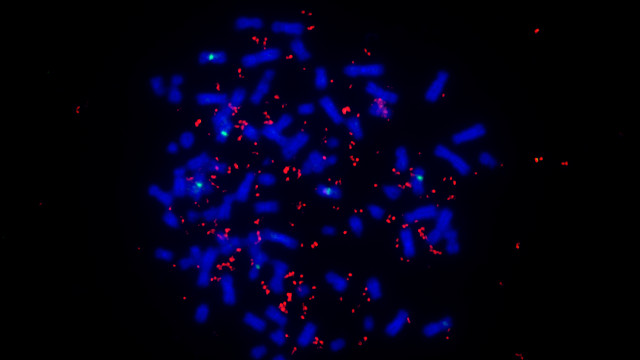
 Metaphase spread of chromosomes (blue) and ecDNA encoding EGFR (red) from a glioblastoma cell line.KRISTEN TURNER, UNIVERSITY OF CALIFORNIA, SAN DIEGOGenetically, tumors are a mess. Individual tumor cells have a unique spectrum of mutations, and this heterogeneity is known to drive tumor evolution and resistance to cancer treatments. Now, in a study published today (February 8) in Nature, Paul Mischel of the Ludwig Institute for Cancer Research at the University of California, San Diego (UCSD), and colleagues report on another level of tumor heterogeneity: widespread non-chromosomal DNA—likely to be circular DNA—found in 40 percent of the tumor cell lines analyzed. This extra-chromosomal DNA (ecDNA) expressed multiple copies of oncogenes associated with drive tumor growth and survival. The finding suggests that this non-chromosomal DNA likely contributes to tumor heterogeneity and evolution.
Metaphase spread of chromosomes (blue) and ecDNA encoding EGFR (red) from a glioblastoma cell line.KRISTEN TURNER, UNIVERSITY OF CALIFORNIA, SAN DIEGOGenetically, tumors are a mess. Individual tumor cells have a unique spectrum of mutations, and this heterogeneity is known to drive tumor evolution and resistance to cancer treatments. Now, in a study published today (February 8) in Nature, Paul Mischel of the Ludwig Institute for Cancer Research at the University of California, San Diego (UCSD), and colleagues report on another level of tumor heterogeneity: widespread non-chromosomal DNA—likely to be circular DNA—found in 40 percent of the tumor cell lines analyzed. This extra-chromosomal DNA (ecDNA) expressed multiple copies of oncogenes associated with drive tumor growth and survival. The finding suggests that this non-chromosomal DNA likely contributes to tumor heterogeneity and evolution.
“This is the first comprehensive study of the frequency of extrachromosomal DNA found in diverse cancer types,” Charles Swanton, who studies cancer diversity at the Francis Crick Institute in the U.K., and who was not involved in the work, wrote in an email to The Scientist. “The results presented illustrate just how prevalent this process is and how important ecDNA is likely to be for altering cancer cell phenotypic behavior to adapt to selection pressures.”
“This study provides yet another layer of dimensionality to how tumors permute their genomic material to become more aggressive and adapt to [anti-cancer] therapy,” said Levi Garraway, head of global oncology development at Eli Lilly, who also was not involved in the work.
Oncogene-encoding ecDNA was thought to be rare ...