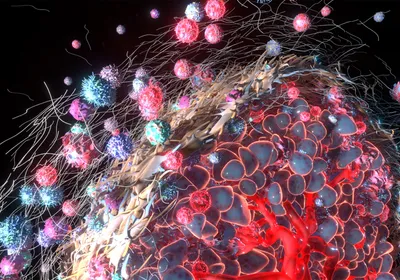
SIFTING STEM CELLS: Gold nanoparticles carrying antibodies to stem cell surface markers bind to the cells and enhance spectral signals detected by surface-enhanced Raman spectroscopy (SERS). J. HAN ET AL., BIOMATERIALS, doi:10.1016/j.biomaterials.2016.07.033, 2016.
SIFTING STEM CELLS: Gold nanoparticles carrying antibodies to stem cell surface markers bind to the cells and enhance spectral signals detected by surface-enhanced Raman spectroscopy (SERS). J. HAN ET AL., BIOMATERIALS, doi:10.1016/j.biomaterials.2016.07.033, 2016.
When a laser beam shines on a single cell, chemicals within the cell can absorb, reflect, or scatter the light waves. The scattered light generates a unique signature at various wavelengths that can be used to identify specific molecules, depending on the molecule type—protein, sugar, or nucleic acid—and the chemical bonds present within its structure. For decades, researchers have used these signatures, known as Raman spectra, as chemical fingerprints to characterize cells.
Unlike flow cytometry, microfluidics, and other cell sorting methods, techniques that rely on Raman spectroscopy do not require labels, and they can be more sensitive and specific than flow cytometry for many applications. Variant versions such as surface-enhanced Raman spectroscopy (SERS)—where chemicals are adsorbed on the surface of a metal, boosting the emitted ...